Chapter 18 NPS_contrast ~ cue * stim
18.1 Overview
The purpose of this markdown is to plot the NPS extracted values for each contrast of interest. Steps of this notebook entails the following:
- load and stack the files
- plot per condition
- x axis (pain vs. vicarious vs. cognitive)
- y axis (NPS extracted values)
# contrast_name = {'P_VC_cue_high_gt_low', 'V_PC_cue_high_gt_low', 'C_PV_cue_high_gt_low', ...
# 'P_VC_stimlin_high_gt_low', 'V_PC_stimlin_high_gt_low', 'C_PV_stimlin_high_gt_low',...
# 'P_VC_stimquad_med_gt_other', 'V_PC_stimquad_med_gt_other', 'C_PV_stimquad_med_gt_other',...
# 'P_VC_cue_int_stimlin','V_PC_cue_int_stimlin', 'C_PV_cue_int_stimlin',...
# 'P_VC_cue_int_stimquad','V_PC_cue_int_stimquad','C_PV_cue_int_stimquad',...
# 'motor',...
# 'P_simple_cue_high_gt_low', 'V_simple_cue_high_gt_low', 'C_simple_cue_high_gt_low', ...
# 'P_simple_stimlin_high_gt_low', 'V_simple_stimlin_high_gt_low', 'C_simple_stimlin_high_gt_low',...
# 'P_simple_stimquad_med_gt_other', 'V_simple_stimquad_med_gt_other', 'C_simple_stimquad_med_gt_other',...
# 'P_simple_cue_int_stimlin', 'V_simple_cue_int_stimlin', 'C_simple_cue_int_stimlin',...
# 'P_simple_cue_int_stimquad','V_simple_cue_int_stimquad','C_simple_cue_int_stimquad'18.2 regressors and contrasts
What regressors were used in the neural model and how did you create contrasts?
This Rmd is based on the univariate analysis mainly using 2 factors (cue x stimulus intensity).
The 6 regressors of interest are
- high-cue_high-stim
- high-cue_med-stim
- high-cue_low-stim
- low-cue_high-stim
- low-cue_med-stim
- low-cue_low-stim.
If interested, the variable of interest is coded “
onset03_stim” in the behavioral data.
Additional regressors include 7) cue_onset “
onset01_cue”, 8) onset of the expectation rating phase “onset02_ratingexpect” convolved with the reaction time of the expectation rating “pmod_expectRT”, and 9) onset of the outcome rating phase “onset04_ratingoutcome”, convolved with the reaction time of the outcome rating “pmod_outcomeRT”.Motion covariates include a) csf, b) 24 DOF head motion variables, and c) spikes derived using a FD-spike-threshold of 0.9mm. Participants with a motion spike of more than 20 per run is excluded from the analysis. For the 6 regressors of interest, I build 5 contrasts that capture the cue effect, the stimulus intensity effect, and the interaction of these two factors.
| contrast names | high-cue_high-stim | high-cue_med-stim | high-cue_low-stim | low-cue_high-stim | low-cue_med-stim | low-cue_low-stim |
|---|---|---|---|---|---|---|
| cue_high_gt_low | 1 | 1 | 1 | -1 | -1 | -1 |
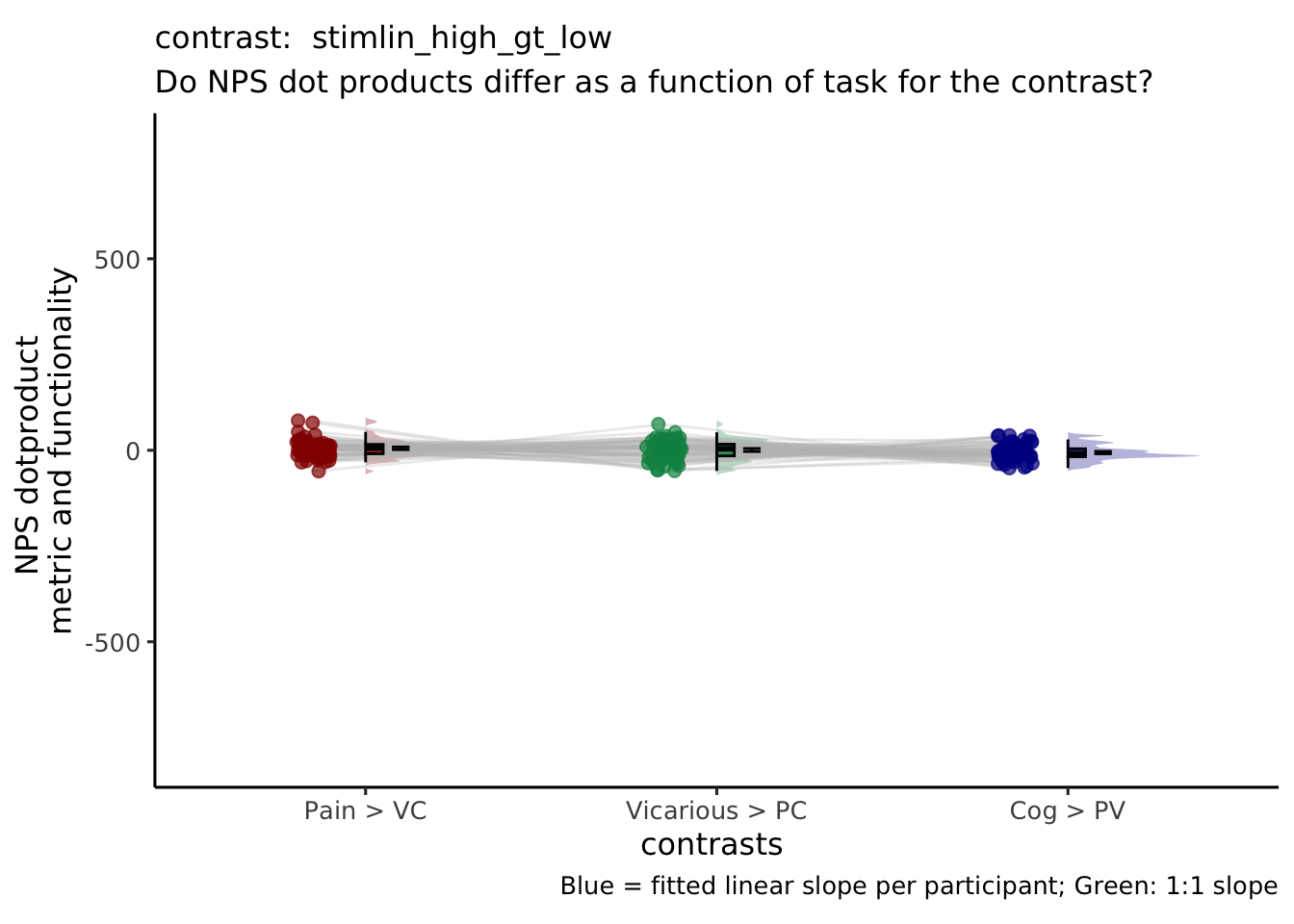
| stimlin_high_gt_low | 1 | 0 | -1 | 1 | 0 | -1 |
| stimquad_med_gt_other | -1 | 2 | -1 | -1 | 2 | -1 |
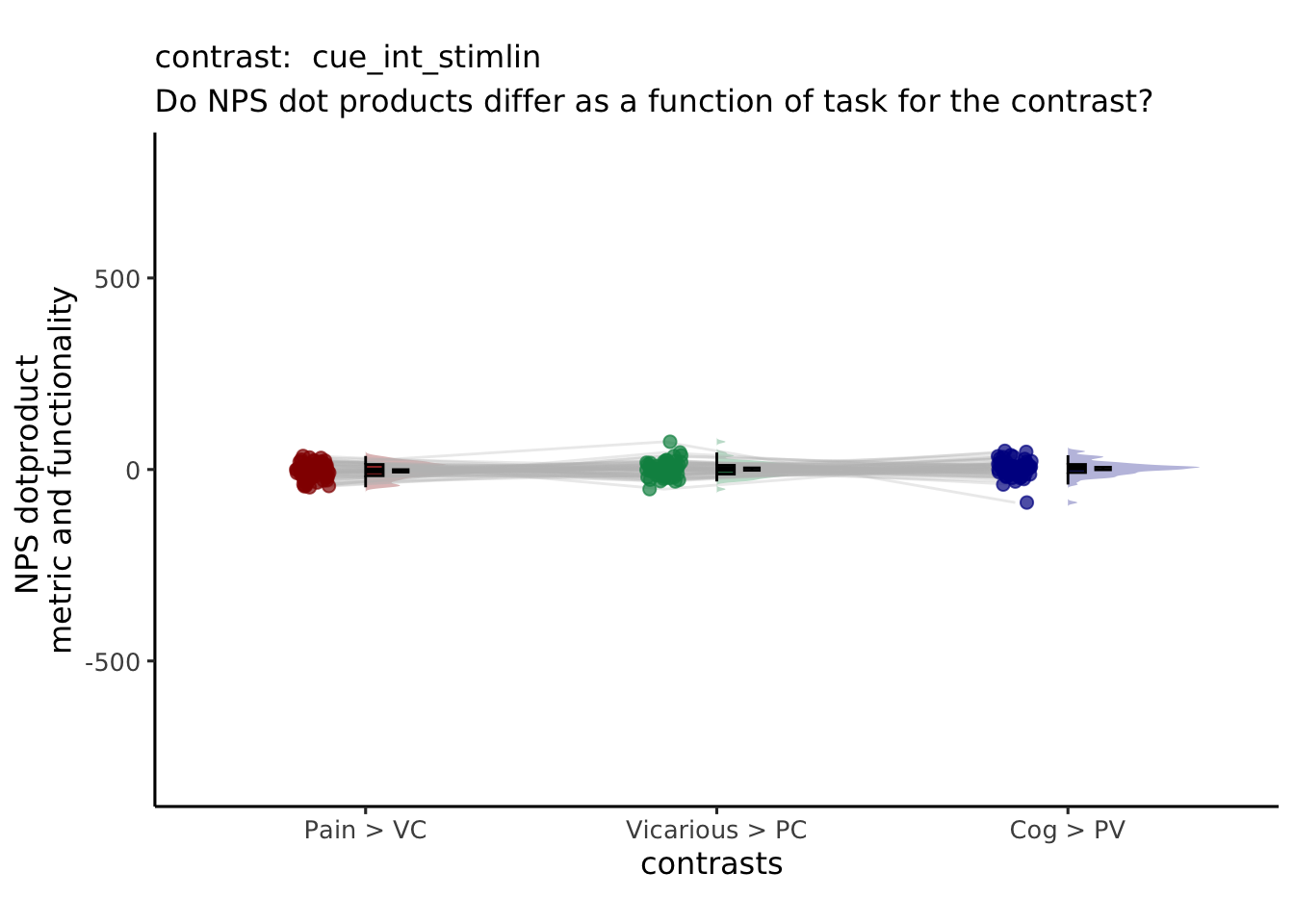
| cue_int_stimlin | 1 | 0 | -1 | -1 | 0 | 1 |
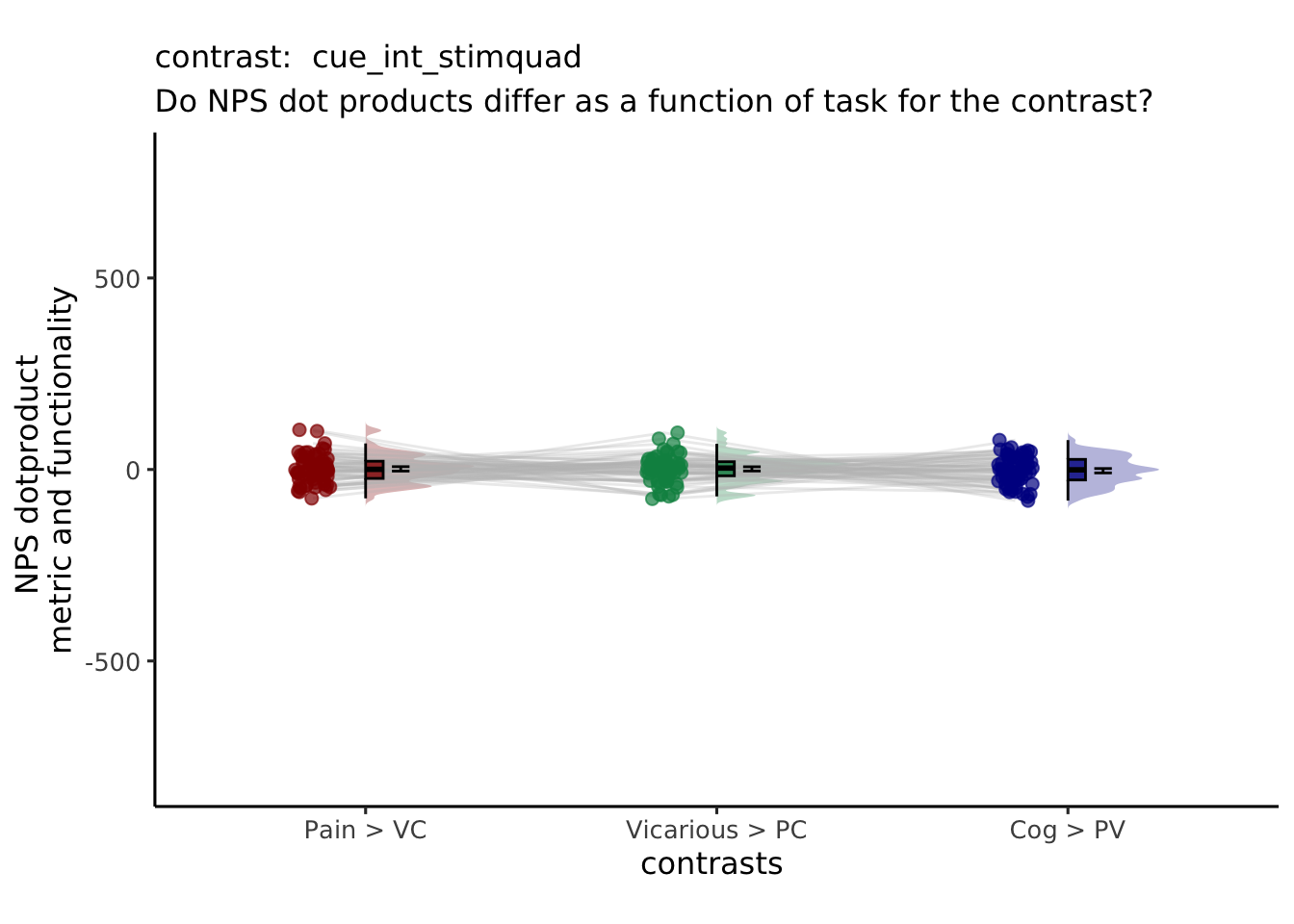
| cue_int_stimquad | -1 | 2 | -1 | 1 | -2 | 1 |
Here are the common parameters that I use for ggplot
npsdir = file.path(main_dir,'analysis','fmri','spm','univariate','model01_6cond','extract_nps')
model = 'nps'; model_keyword = "nps"
subjectwise_mean = "mean_per_sub"; group_mean = "mean_per_sub_norm_mean"; se = "se"
iv = "contrast"; subject = "subject"
dv = "nps"; dv_keyword = "nps_dot_product"
ylim = c(-800, 800)
xlab = "contrasts "; ylab = "NPS dotproduct \nmetric and functionality"
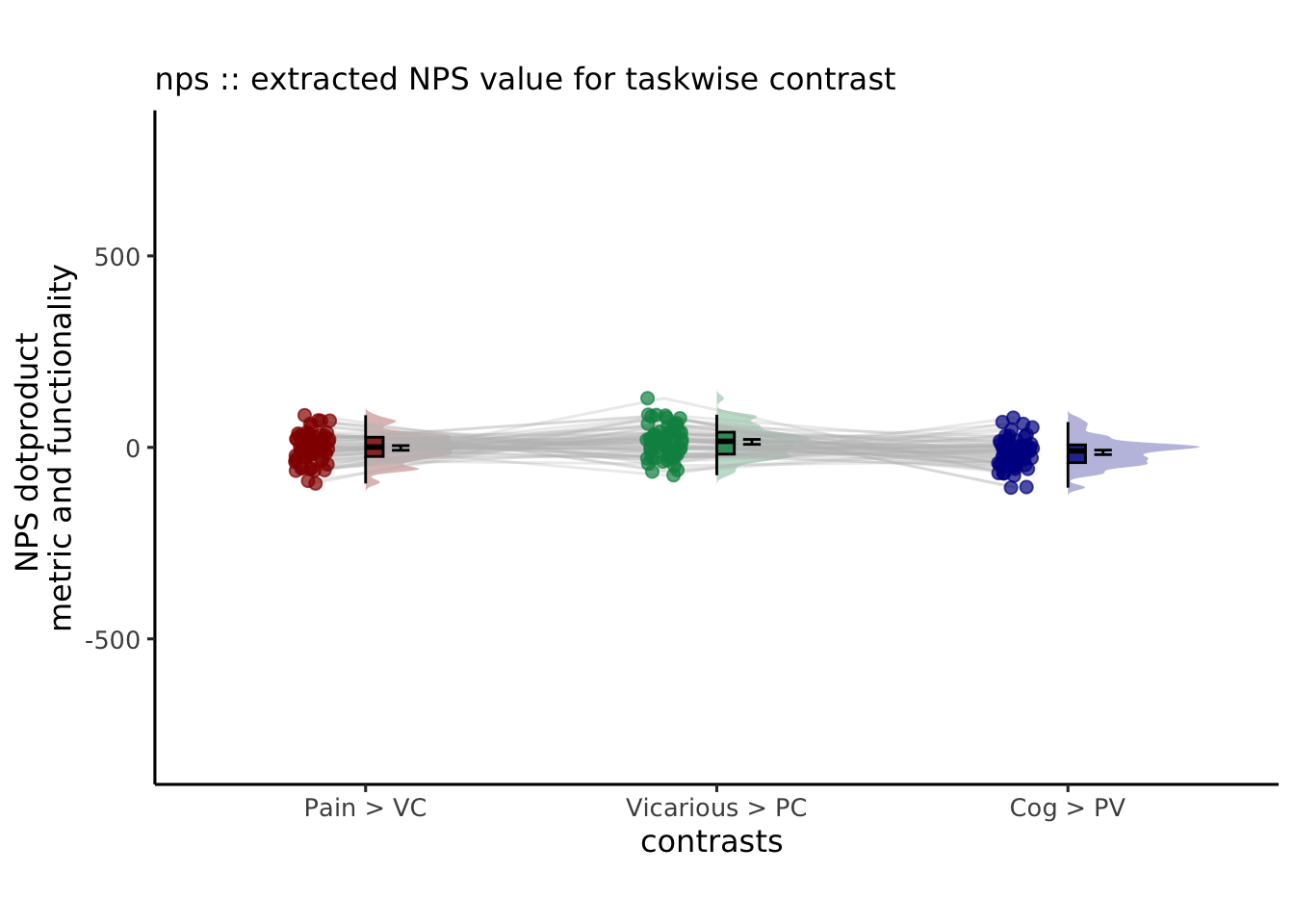
ggtitle = paste0(model_keyword,
" :: extracted NPS value for taskwise contrast")
legend_title = "Contrasts"
color_scheme <- c("Pain > VC" = "#941100",
"Vicarious > PC" = "#008F51",
"Cog > PV" = "#011891")
savedir = file.path(main_dir,'analysis','mixedeffect','model10_iv-cue-stim_dv-nps-contrast',as.character(Sys.Date()) )