18 fMRI :: FIR ~ task TTL1
The purpose of this notebook is to plot the BOLD timeseries from SPM FIR model. TODO
- load tsv
- concatenate
- per time column, calculate mean and variance
- plot
18.1 references
https://stackoverflow.com/questions/29402528/append-data-frames-together-in-a-for-loop/29419402
plot_timeseries_onefactor <-
function(df, iv1, mean, error, xlab, ylab, ggtitle, color) {
n_points <- 100 # Number of points for interpolation
g <- ggplot(
data = df,
aes(
x = .data[[iv1]],
y = .data[[mean]],
group = 1,
color = color
),
cex.lab = 1.5,
cex.axis = 2,
cex.main = 1.5,
cex.sub = 1.5
) +
geom_errorbar(aes(
ymin = (.data[[mean]] - .data[[error]]),
ymax = (.data[[mean]] + .data[[error]]),
color = color
),
width = .1,
alpha = 0.8) +
geom_line() +
geom_point(color = color) +
ggtitle(ggtitle) +
xlab(xlab) +
ylab(ylab) +
theme_classic() +
theme(aspect.ratio = .6) +
expand_limits(x = 3.25) +
scale_color_manual("",
values = color) +
# theme(
# legend.position = c(.99, .99),
# legend.justification = c("right", "top"),
# legend.box.just = "right",
# legend.margin = margin(6, 6, 6, 6)
# ) +
# theme(legend.key = element_rect(fill = "white", colour = "white")) +
theme_bw()
return(g)
}
plot_timeseries_bar_SANDBOX <-
function(df, iv1, iv2, mean, error, xlab, ylab, ggtitle, color) {
n_points <- 100 # Number of points for interpolation
## Removing "tr" from the column values
df[[iv1]] <- as.numeric(sub("tr", "", df[[iv1]]))
g <- ggplot(
data = df,
aes(
x = .data[[iv1]],
y = .data[[mean]],
group = factor(.data[[iv2]]),
color = factor(.data[[iv2]])
),
cex.lab = 1.5,
cex.axis = 2,
cex.main = 1.5,
cex.sub = 1.5
) +
geom_errorbar(aes(
ymin = (.data[[mean]] - .data[[error]]),
ymax = (.data[[mean]] + .data[[error]]),
fill = factor(.data[[iv2]])
),
width = .1,
alpha = 0.8) +
geom_line() +
geom_point() +
ggtitle(ggtitle) +
xlab(xlab) +
ylab(ylab) +
theme_classic() +
expand_limits(x = 3.25) +
scale_color_manual("",
values = color) +
scale_fill_manual("",
values = color) +
theme(
aspect.ratio = .6,
text = element_text(size = 20),
axis.title.x = element_text(size = 24),
axis.title.y = element_text(size = 24),
legend.position = c(.99, .99),
legend.justification = c("right", "top"),
legend.box.just = "right",
legend.margin = margin(6, 6, 6, 6)
) +
theme(legend.key = element_rect(fill = "white", colour = "white")) +
theme_bw()
return(g)
}parameters
main_dir <- dirname(dirname(getwd()))
datadir <- file.path(main_dir, "analysis/fmri/nilearn/glm/fir")
analysis_folder <- paste0("model52_iv-6cond_dv-firglasserSPM_ttl1")
analysis_dir <-
file.path(main_dir,
"analysis",
"mixedeffect",
analysis_folder,
as.character(Sys.Date()))
dir.create(analysis_dir,
showWarnings = FALSE,
recursive = TRUE)
save_dir <- analysis_dir18.2 taskwise stim effect
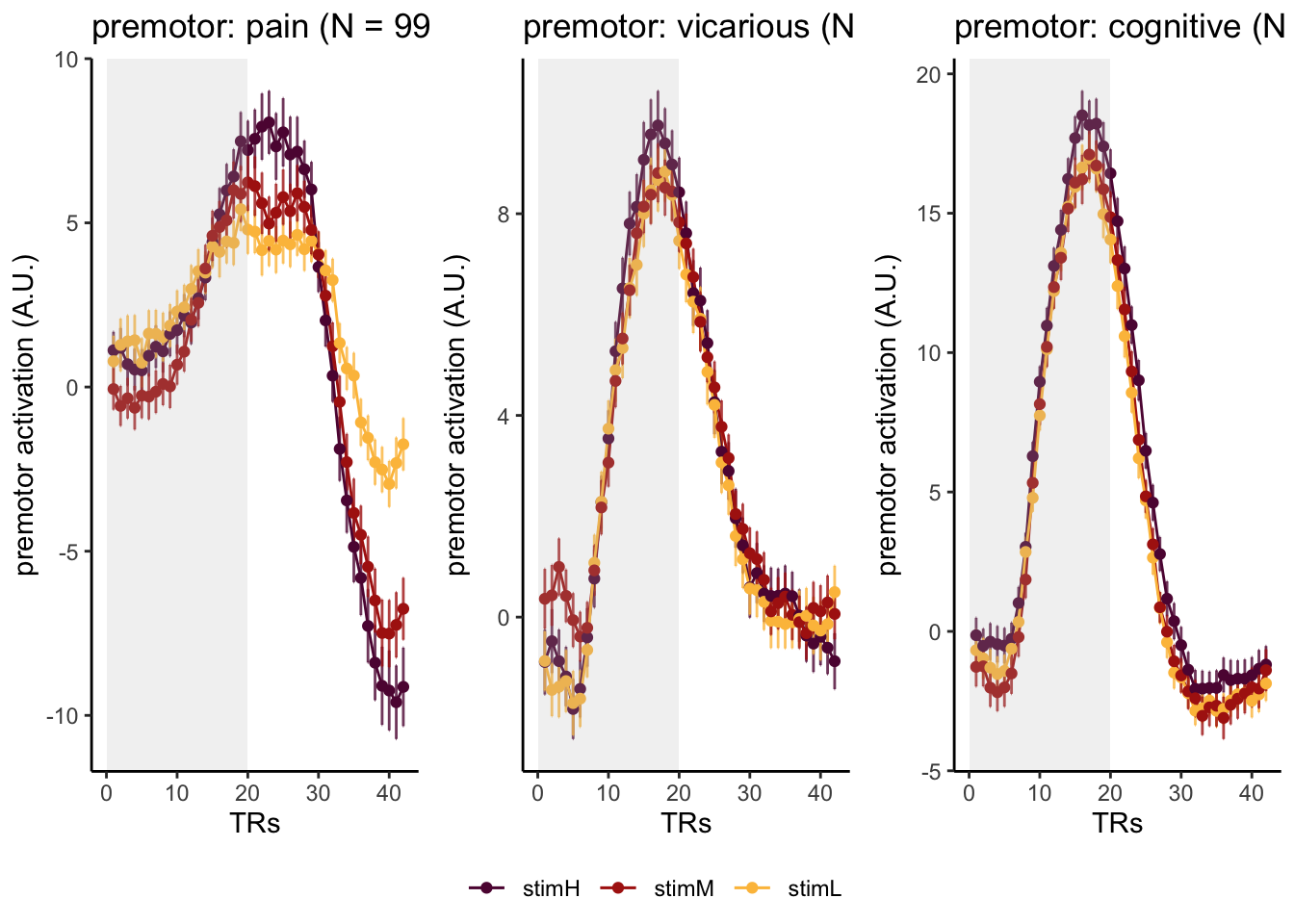
Here, I have a list of ROIs. Per ROI, I have FIR values for pain, vicarious, cognitive tasks. We’ll aggregate data per ROI and plot the time series for the 3 tasks.
roi_list <- c("dACC", "PHG", "V1", "SM", "MT", "RSC", "LOC", "FFC", "PIT", "pSTS", "AIP", "premotor") # 'rINS', 'TPJ',
run_types <- c("pain", "vicarious", "cognitive")
plot_list <- list()
TR_length <- 42
for (ROI in roi_list) {
main_dir <- dirname(dirname(getwd()))
datadir <- file.path(main_dir, "analysis/fmri/spm/fir/ttl1par")
taskname <- "pain"
exclude <- "sub-0001"
filename <- paste0("sub-*", "*roi-", ROI, "_tr-42.csv")
common_path <- Sys.glob(file.path(datadir, "sub-*", filename))
filter_path <-
common_path[!str_detect(common_path, pattern = exclude)]
df <-
do.call("rbind.fill", lapply(
filter_path,
FUN = function(files) {
read.table(files, header = TRUE, sep = ",")
}
))
for (run_type in run_types) {
print(run_type)
filtered_df <-
df[!(df$condition == "rating" |
df$condition == "cue" | df$runtype != run_type),]
parsed_df <- filtered_df %>%
separate(
condition,
into = c("cue", "stim"),
sep = "_",
remove = FALSE
)
# --------------------------------------------------------------------------
# 0) subset dataframe based on ROI
# --------------------------------------------------------------------------
df_long <-
pivot_longer(
parsed_df,
cols = starts_with("tr"),
names_to = "tr_num",
values_to = "tr_value"
)
# --------------------------------------------------------------------------
# 1) clean factor
# --------------------------------------------------------------------------
df_long$tr_ordered <- factor(df_long$tr_num,
levels = c(paste0("tr", 1:TR_length)))
df_long$stim_ordered <- factor(df_long$stim,
levels = c("stimH", "stimM", "stimL"))
# --------------------------------------------------------------------------
# 2) summary statistics
# --------------------------------------------------------------------------
subjectwise <- meanSummary(df_long,
c("sub", "tr_ordered", "stim_ordered"),
"tr_value")
groupwise <- summarySEwithin(
data = subjectwise,
measurevar = "mean_per_sub",
withinvars = c("stim_ordered", "tr_ordered"),
idvar = "sub"
)
groupwise$task <- run_type
# https://stackoverflow.com/questions/29402528/append-data-frames-together-in-a-for-loop/29419402
LINEIV1 <- "tr_ordered"
LINEIV2 <- "stim_ordered"
MEAN <- "mean_per_sub_norm_mean"
ERROR <- "se"
dv_keyword <- "actual"
sorted_indices <- order(groupwise$tr_ordered)
groupwise_sorted <- groupwise[sorted_indices,]
# --------------------------------------------------------------------------
# 3) plot per run
# --------------------------------------------------------------------------
p1 <- plot_timeseries_bar_SANDBOX(
groupwise_sorted,
LINEIV1,
LINEIV2,
MEAN,
ERROR,
xlab = "TRs",
ylab = paste0(ROI, " activation (A.U.)"),
ggtitle = paste0(
ROI,
": ",
run_type,
" (N = ",
length(unique(subjectwise$sub)),
") time series, Epoch - stimulus"
),
color = c("#5f0f40", "#ae2012", "#fcbf49")
)
time_points <- seq(1, 0.46 * TR_length, 0.46)
p1 <- p1 +
annotate(
"rect",
xmin = 0,
xmax = 20,
ymin = min(df[[MEAN]], na.rm = TRUE) - 5,
ymax = max(df[[MEAN]], na.rm = TRUE) + 5,
fill = "grey",
alpha = 0.2
)
plot_list[[run_type]] <- p1 + theme_classic()
}
# --------------------------------------------------------------------------
# 4) plot three tasks per ROI
# --------------------------------------------------------------------------
library(gridExtra)
plot_list <- lapply(plot_list, function(plot) {
plot + theme(plot.margin = margin(5, 5, 5, 5)) # Adjust plot margins if needed
})
combined_plot <- ggpubr::ggarrange(
plot_list[["pain"]],
plot_list[["vicarious"]],
plot_list[["cognitive"]],
common.legend = TRUE,
legend = "bottom",
ncol = 3,
nrow = 1,
widths = c(3, 3, 3),
heights = c(.5, .5, .5),
align = "v"
)
print(combined_plot)
ggsave(file.path(
save_dir,
paste0("roi-", ROI, "_epoch-stim_desc-highstimGTlowstim.png")
),
combined_plot,
width = 12,
height = 4)
}## [1] "pain"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "vicarious"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "cognitive"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf##
## Attaching package: 'gridExtra'## The following object is masked from 'package:dplyr':
##
## combine## [1] "pain"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "vicarious"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "cognitive"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf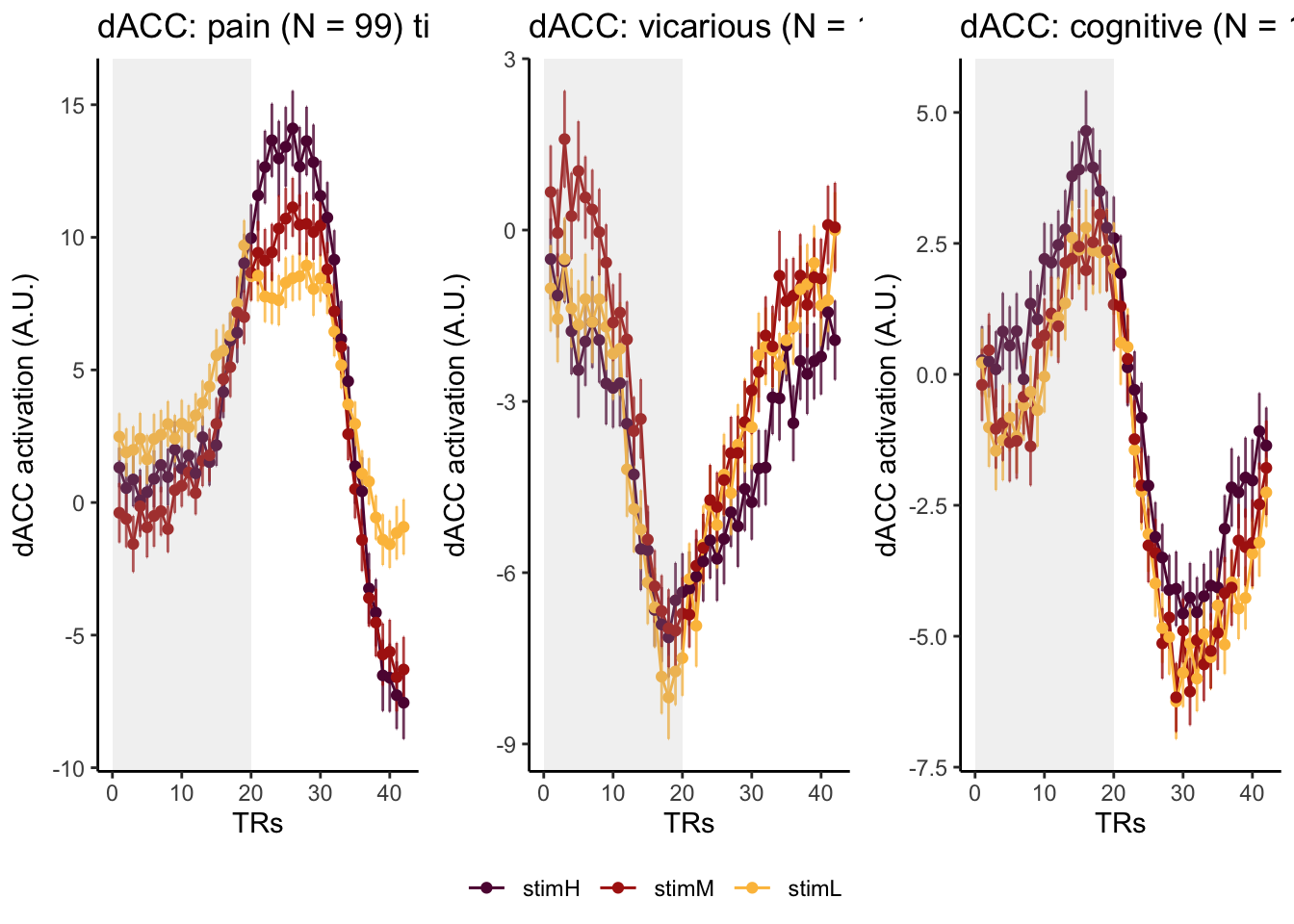
## [1] "pain"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "vicarious"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "cognitive"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf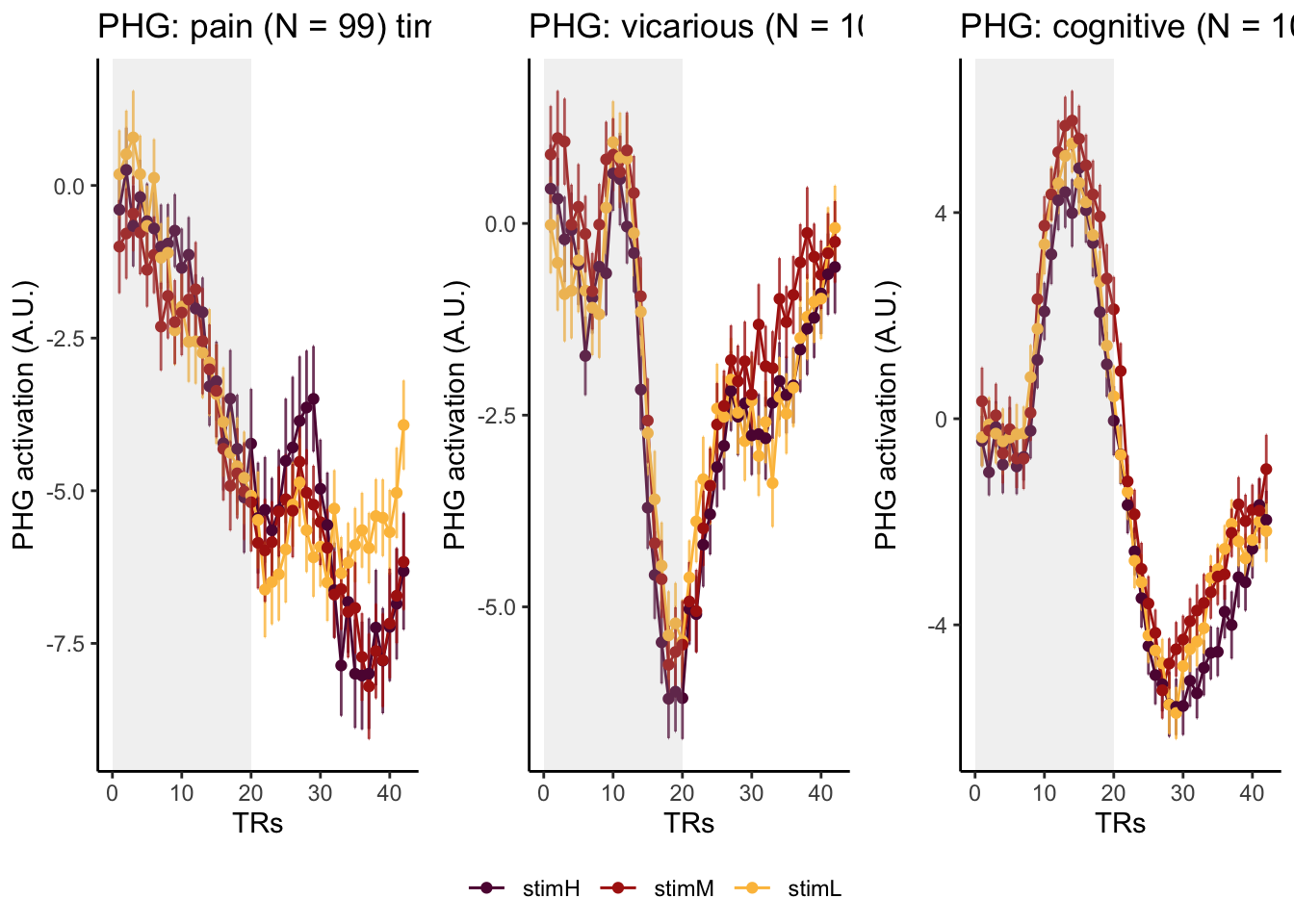
## [1] "pain"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "vicarious"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "cognitive"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf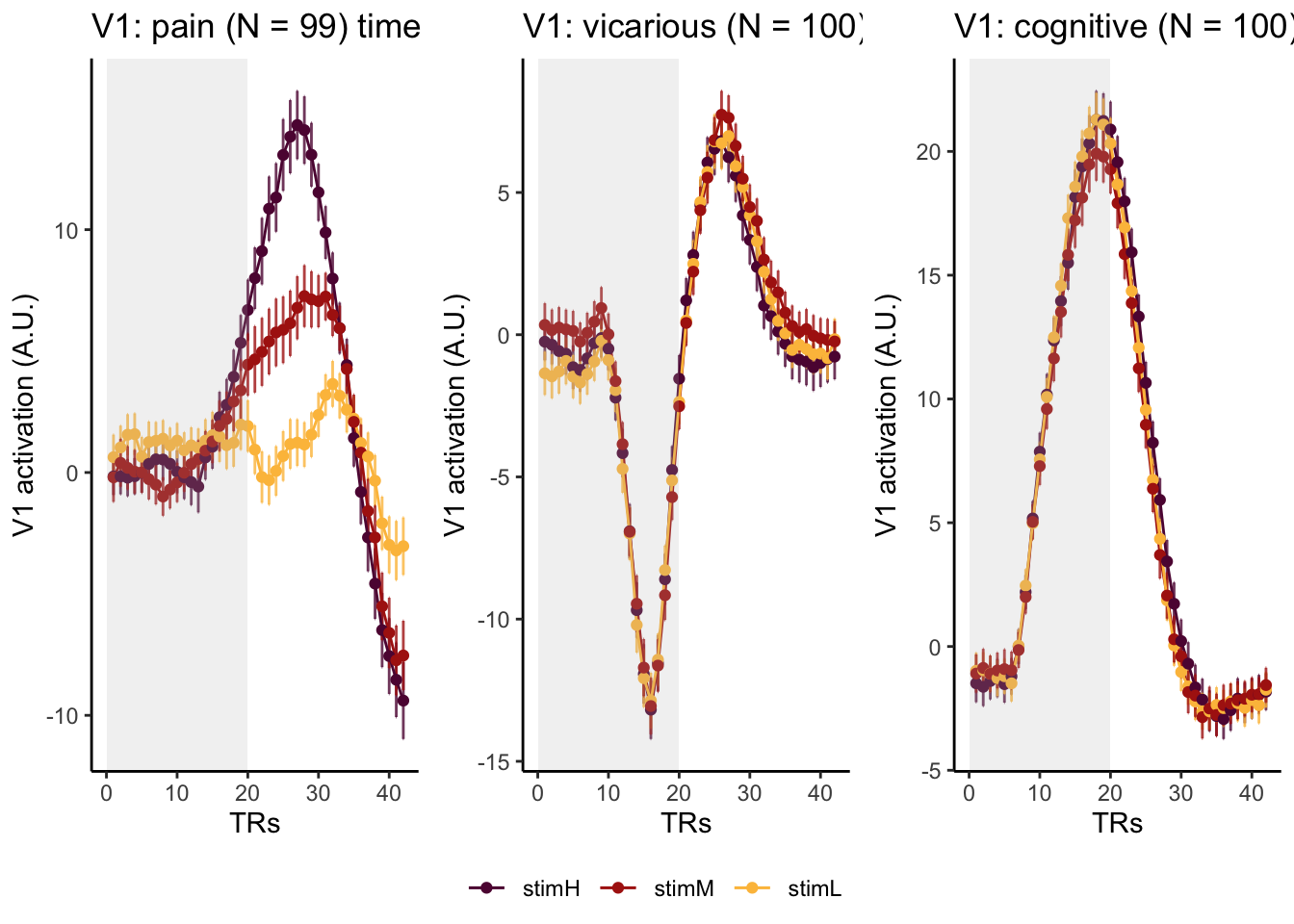
## [1] "pain"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "vicarious"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "cognitive"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf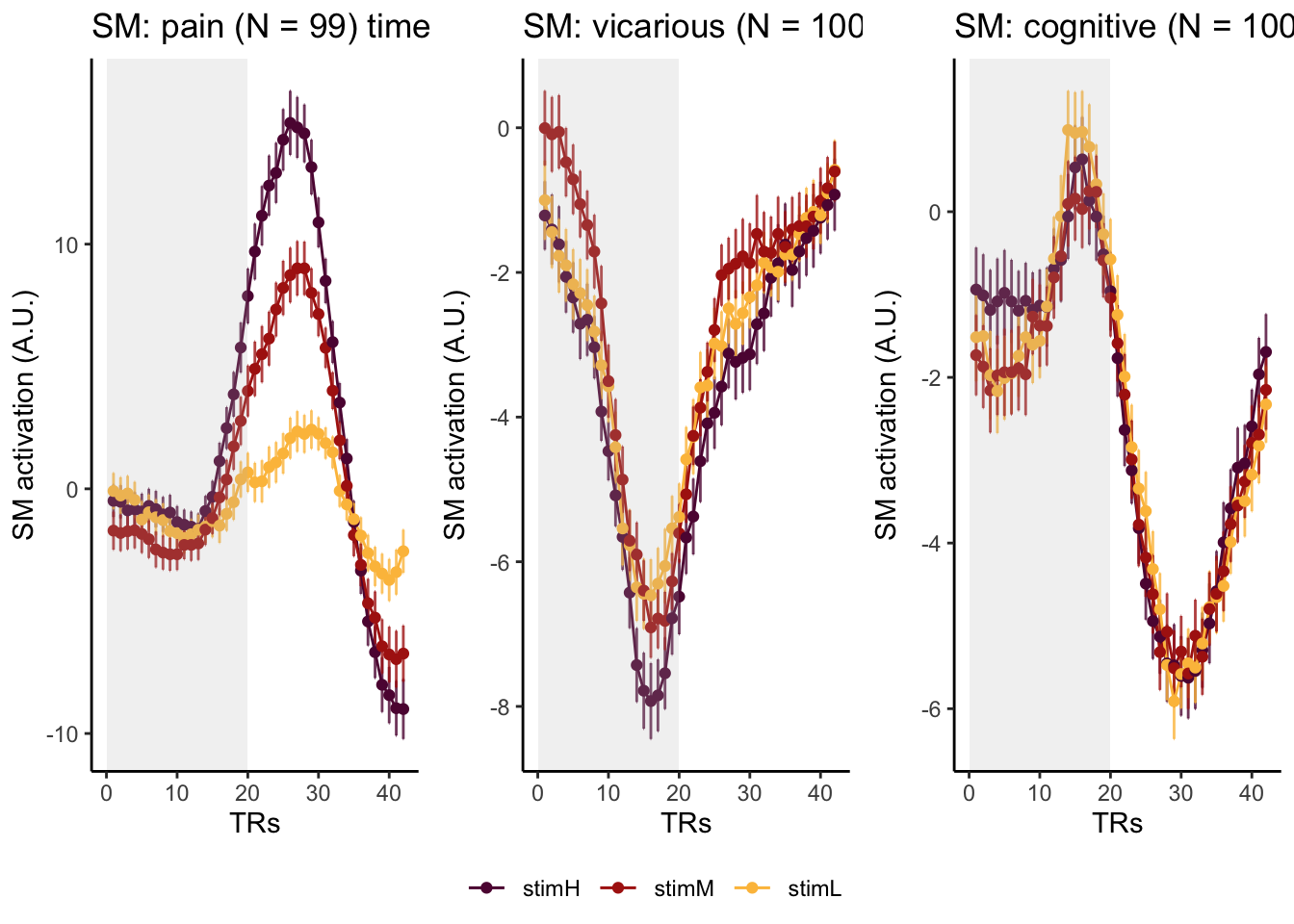
## [1] "pain"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "vicarious"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "cognitive"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf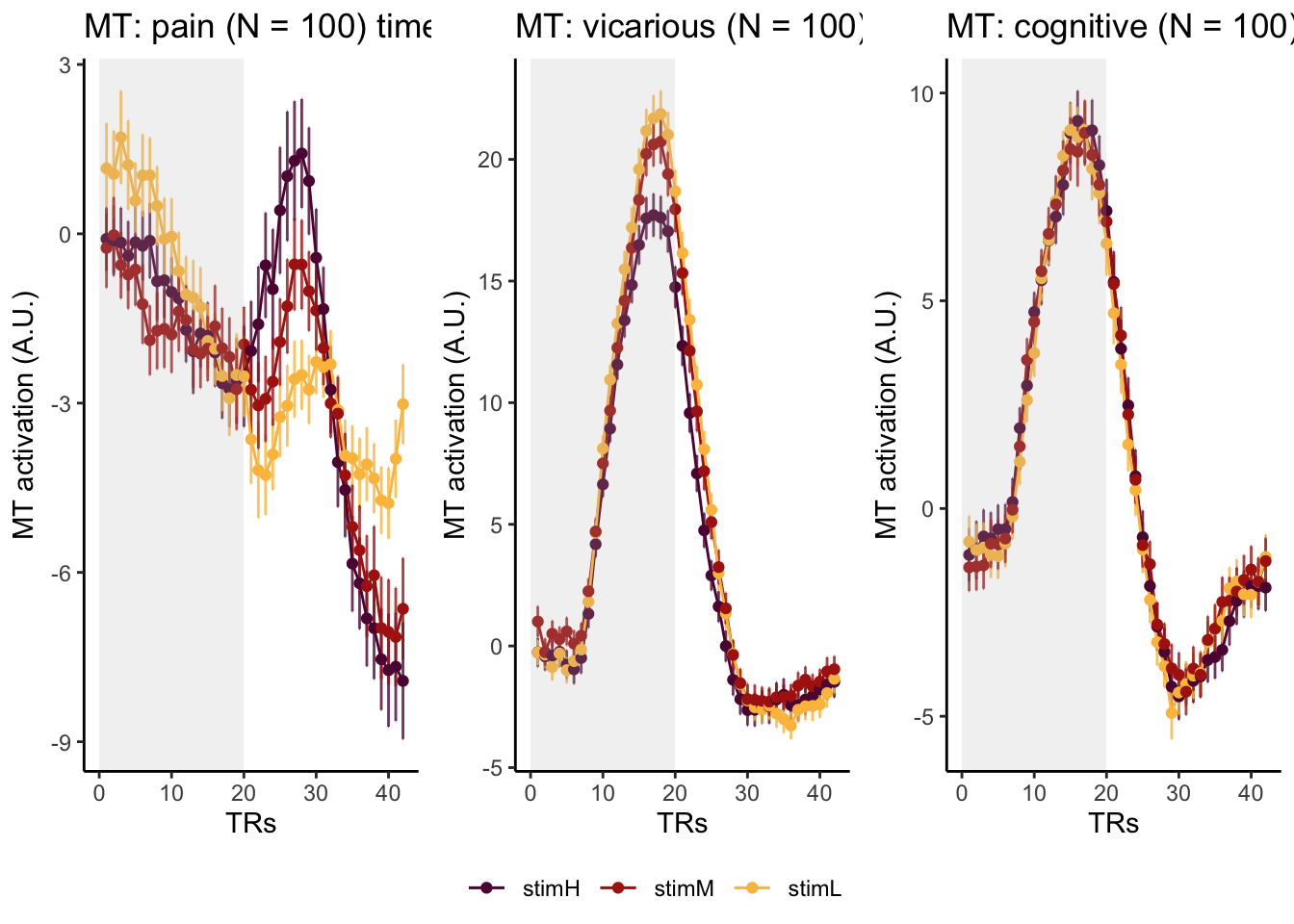
## [1] "pain"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "vicarious"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "cognitive"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf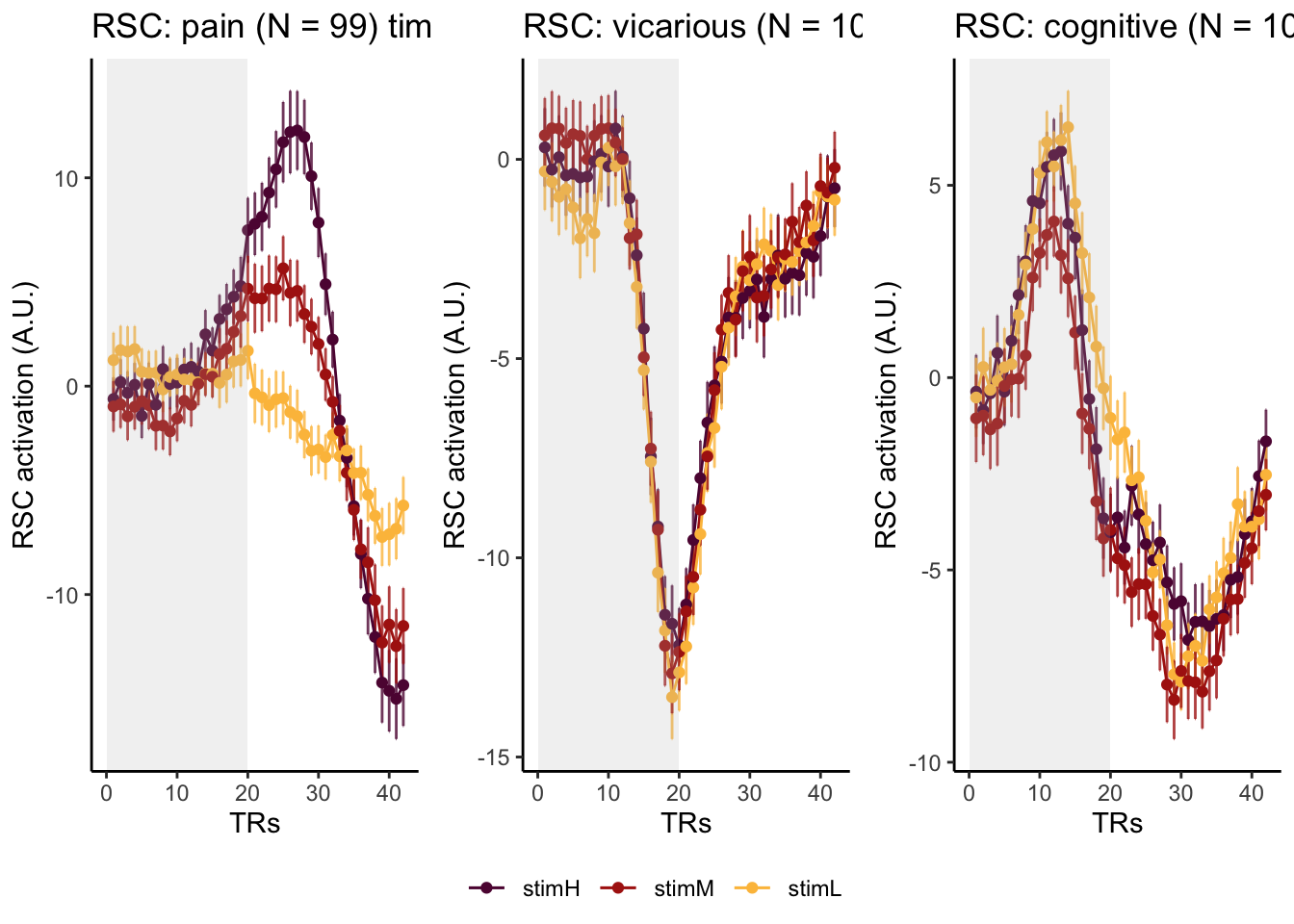
## [1] "pain"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "vicarious"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "cognitive"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf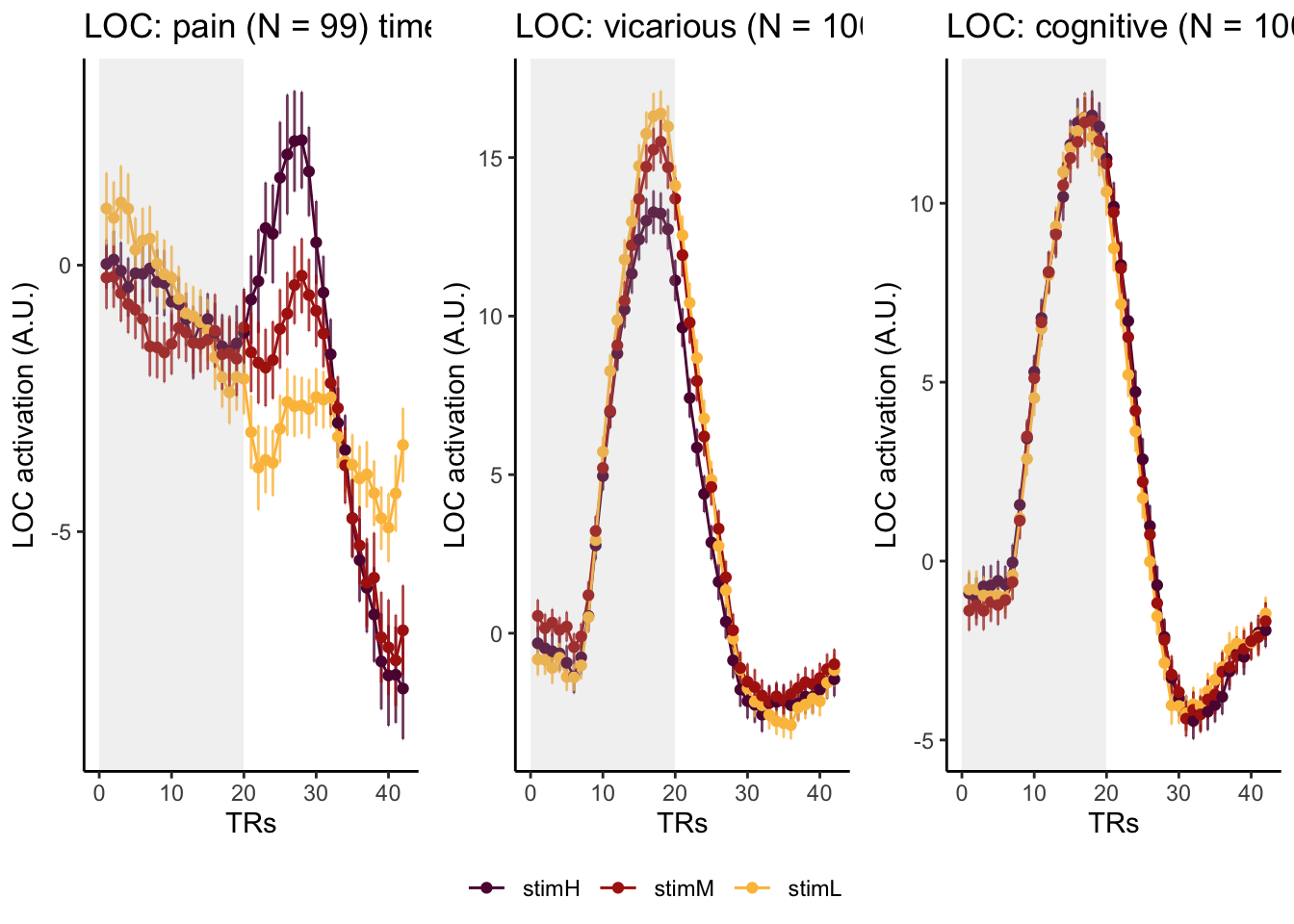
## [1] "pain"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "vicarious"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "cognitive"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf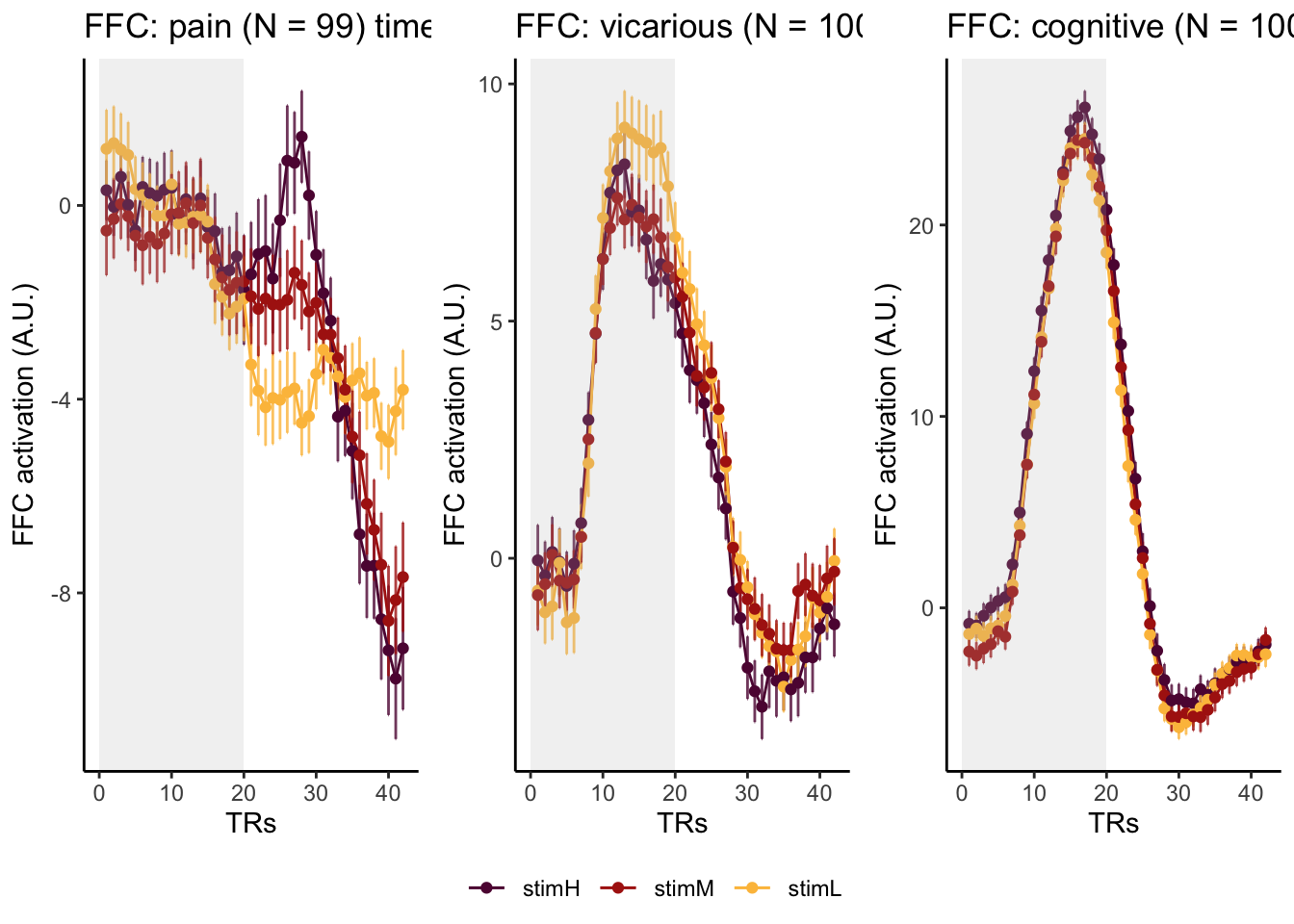
## [1] "pain"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "vicarious"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "cognitive"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf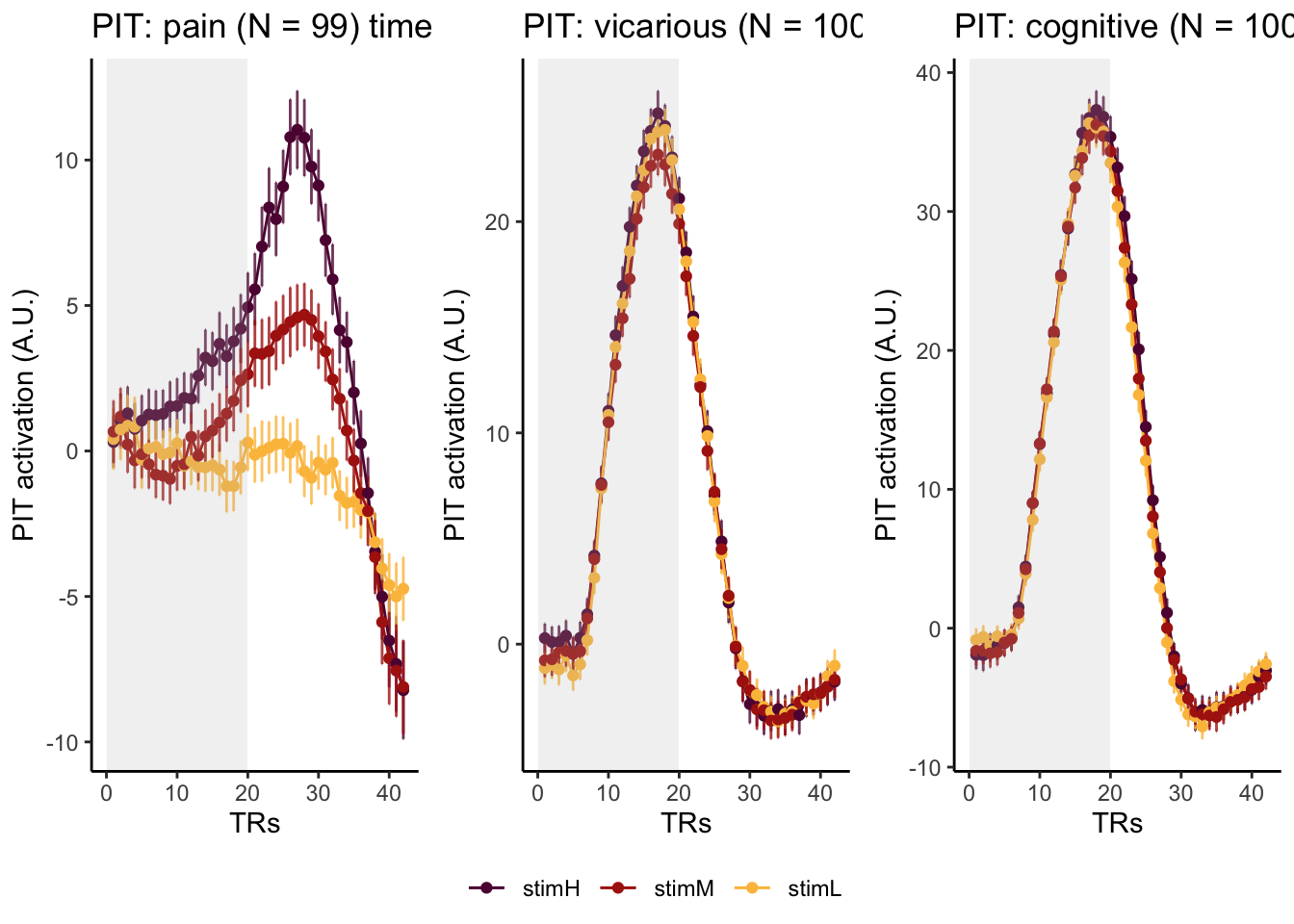
## [1] "pain"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "vicarious"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "cognitive"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf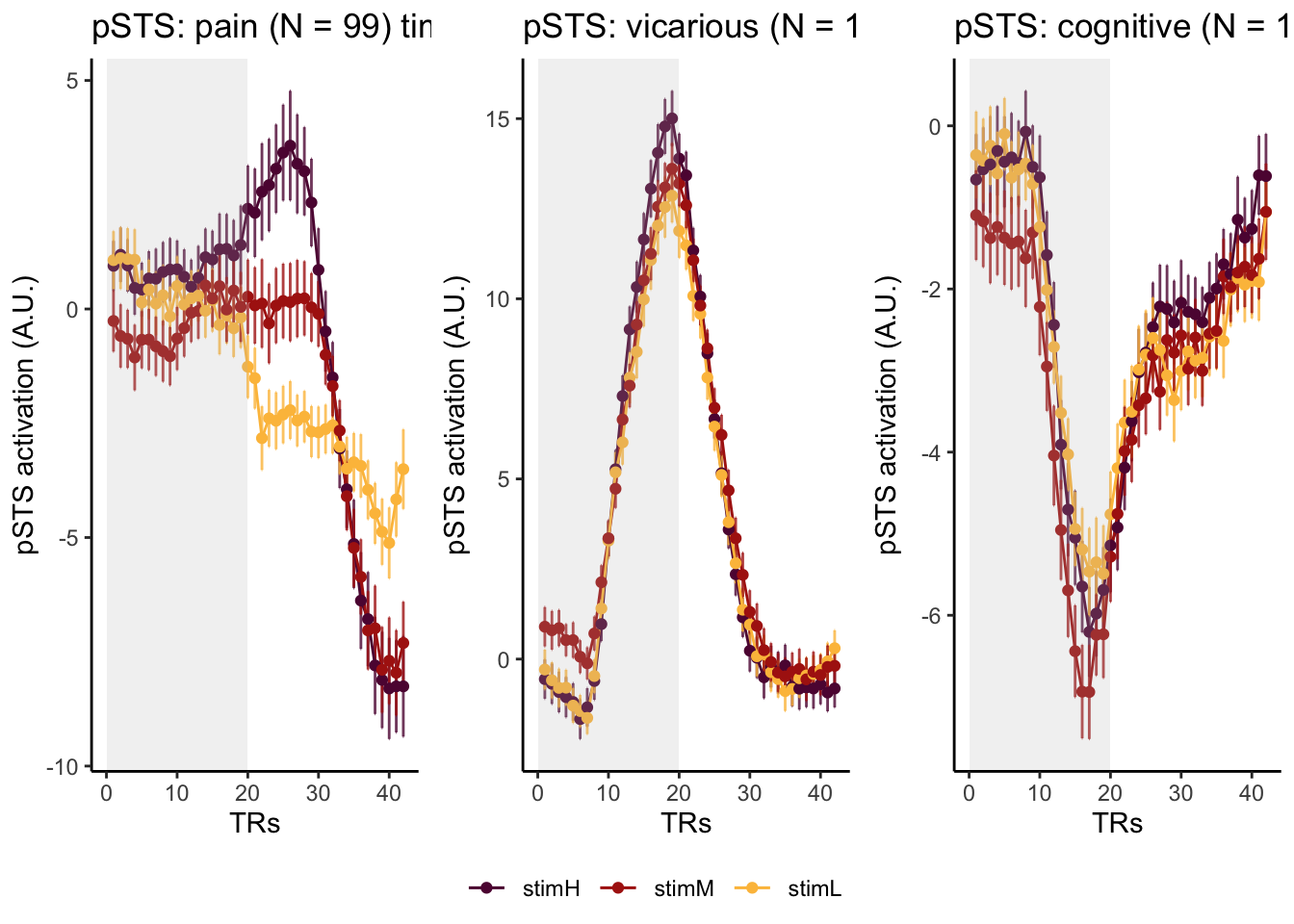
## [1] "pain"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "vicarious"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf## [1] "cognitive"## Warning in geom_errorbar(aes(ymin = (.data[[mean]] - .data[[error]]), ymax =
## (.data[[mean]] + : Ignoring unknown aesthetics: fill## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf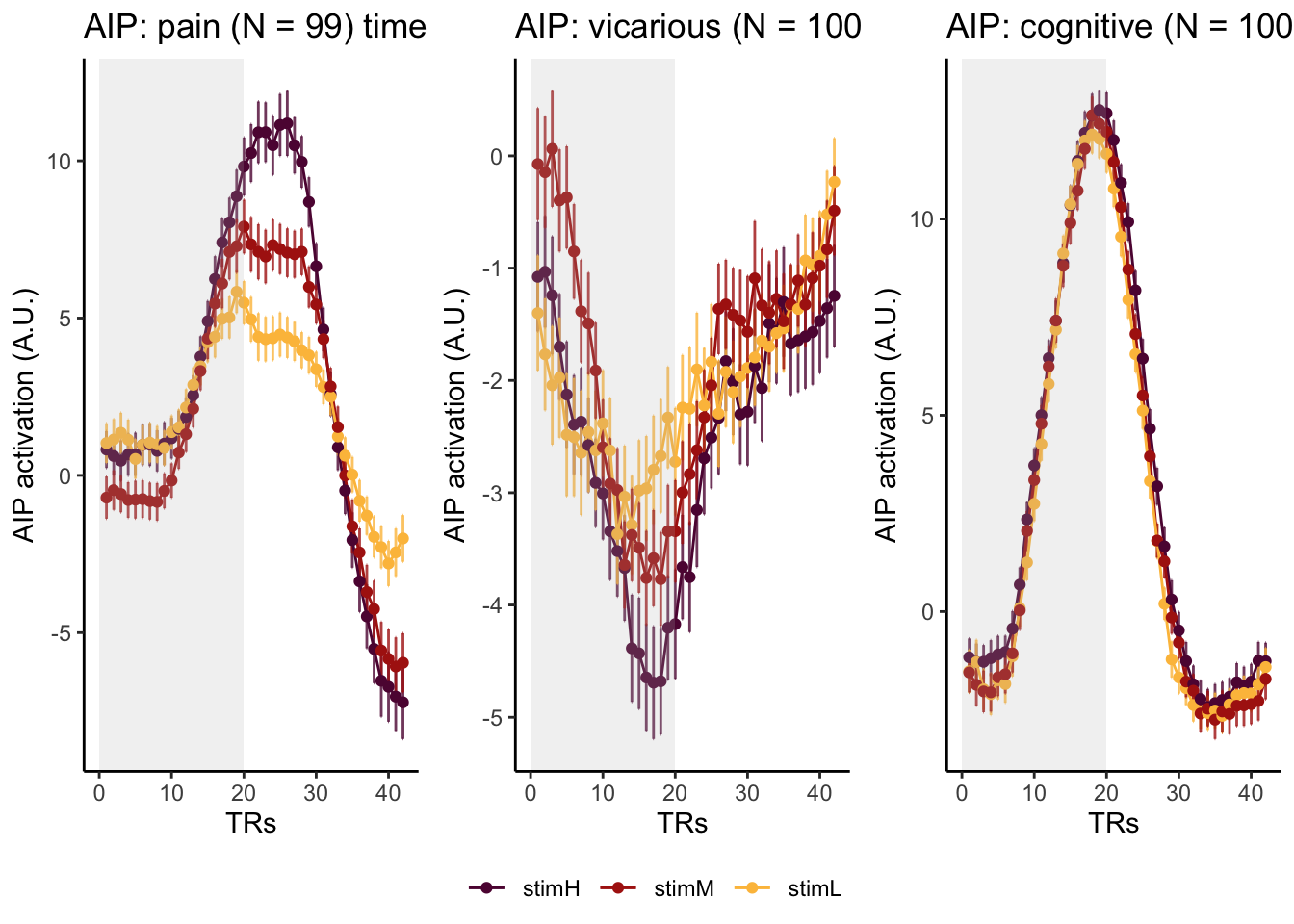
p1 + annotate("rect", xmin = 0, xmax = 10, ymin = min(df[[MEAN]], na.rm = TRUE) - 5, ymax = max(df[[MEAN]], na.rm = TRUE) + 5, fill = "grey", alpha = 0.2)## Warning in min(df[[MEAN]], na.rm = TRUE): no non-missing arguments to min;
## returning Inf## Warning in max(df[[MEAN]], na.rm = TRUE): no non-missing arguments to max;
## returning -Inf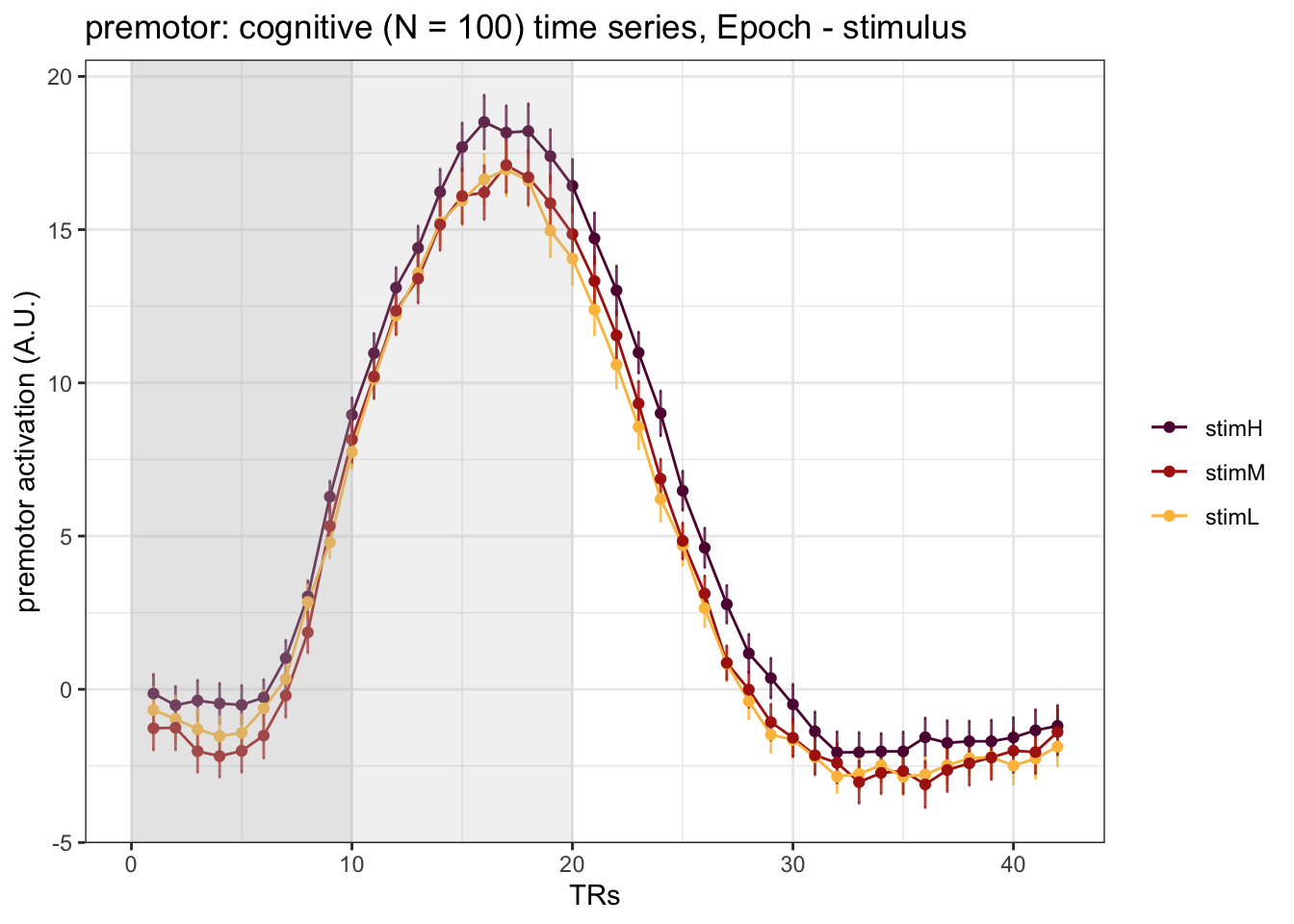
18.2.1 PCA subjectwise
run_types <- c("pain")
for (run_type in run_types) {
print(run_type)
filtered_df <-
df[!(df$condition == "rating" |
df$condition == "cue" | df$runtype != run_type),]
parsed_df <- filtered_df %>%
separate(
condition,
into = c("cue", "stim"),
sep = "_",
remove = FALSE
)
# ------------------------------------------------------------------------------
# subset regions based on ROI
# ------------------------------------------------------------------------------
df_long <-
pivot_longer(
parsed_df,
cols = starts_with("tr"),
names_to = "tr_num",
values_to = "tr_value"
)
# ------------------------------------------------------------------------------
# clean factor
# ------------------------------------------------------------------------------
df_long$tr_ordered <- factor(df_long$tr_num,
levels = c(paste0("tr", 1:TR_length)))
df_long$stim_ordered <- factor(df_long$stim,
levels = c("stimH", "stimM", "stimL"))
# ------------------------------------------------------------------------------
# summary stats
# ------------------------------------------------------------------------------
subjectwise <- meanSummary(df_long,
c("sub", "tr_ordered", "stim_ordered"), "tr_value")
# ------------------------------------------------------------------------------
# convert dataframe long to wide
# ------------------------------------------------------------------------------
df_wide <- pivot_wider(
subjectwise,
id_cols = c("tr_ordered", "stim_ordered"),
names_from = c("sub"),
values_from = "mean_per_sub"
)
stim_high.df <- df_wide[df_wide$stim_ordered == "stimH",]
stim_med.df <- df_wide[df_wide$stim_ordered == "stimM",]
stim_low.df <- df_wide[df_wide$stim_ordered == "stimL",]
meanhighdf <-
data.frame(subset(stim_high.df, select = 3:(ncol(stim_high.df) - 1)))
high.pca_result <- prcomp(meanhighdf)
high.pca_scores <- as.data.frame(high.pca_result$x)
# Access the proportion of variance explained by each principal component
high.variance_explained <-
high.pca_result$sdev ^ 2 / sum(high.pca_result$sdev ^ 2)
plot(high.variance_explained)
# Access the standard deviations of each principal component
high.stdev <- high.pca_result$sdev
meanmeddf <-
data.frame(subset(stim_med.df, select = 3:(ncol(stim_med.df) - 1)))
med.pca <- prcomp(meanmeddf)
med.pca_scores <- as.data.frame(med.pca$x)
meanlowdf <-
data.frame(subset(stim_low.df, select = 3:(ncol(stim_low.df) - 1)))
low.pca <- prcomp(meanlowdf)
low.pca_scores <- as.data.frame(low.pca$x)
combined_pca_scores <-
rbind(high.pca_scores, med.pca_scores, low.pca_scores)
# Add a new column to indicate the stim_ordered category (high_stim or low_stim)
combined_pca_scores$stim_ordered <- c(rep("high_stim", nrow(high.pca_scores)),
rep("med_stim", nrow(med.pca_scores)),
rep("low_stim", nrow(low.pca_scores)))
# ------------------------------------------------------------------------------
# 3d PCA plot
# ------------------------------------------------------------------------------
plot_ly(
combined_pca_scores,
x = ~ PC1,
y = ~ PC2,
z = ~ PC3,
type = "scatter3d",
mode = "markers",
color = ~ stim_ordered
)
}## [1] "pain"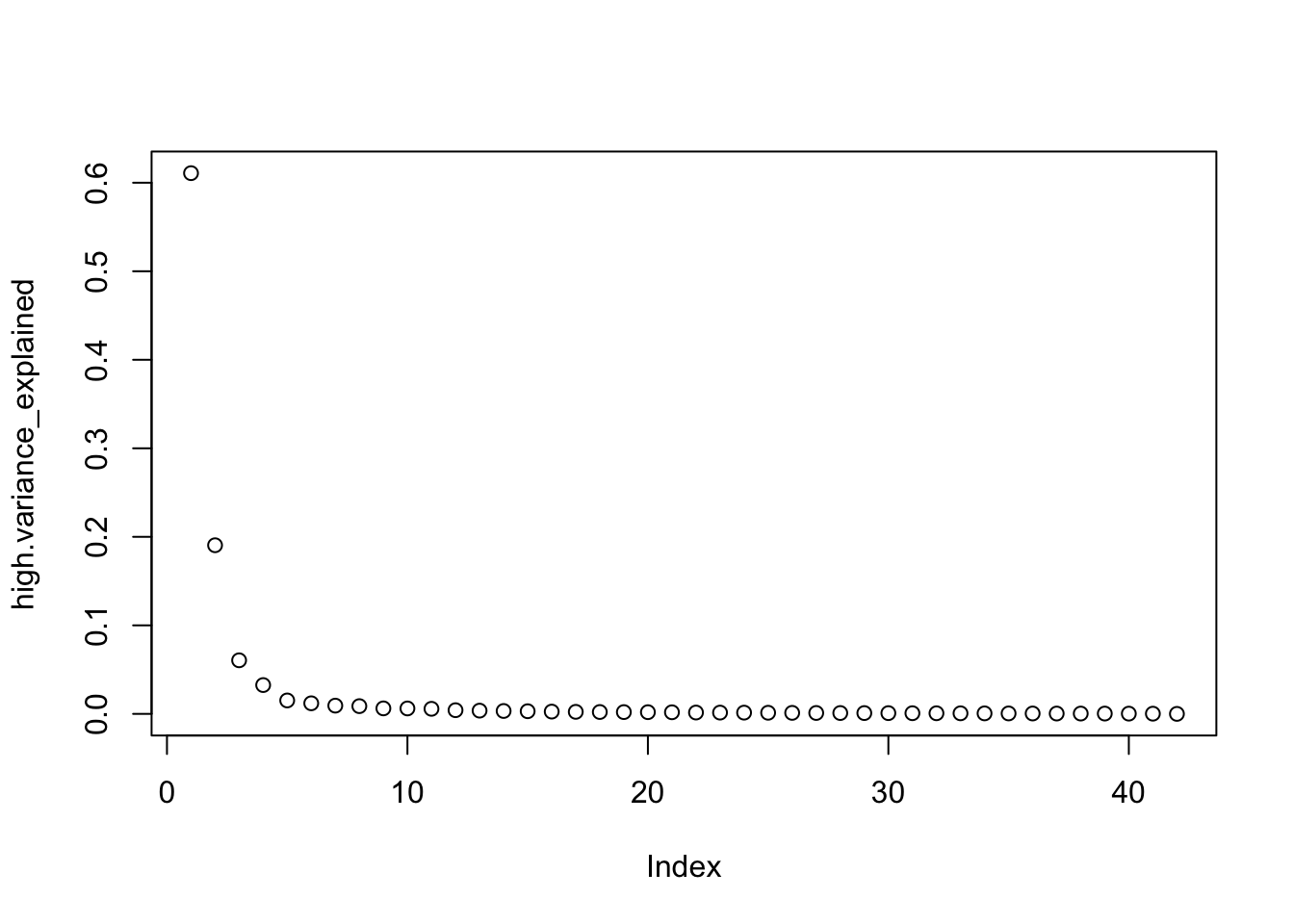
plot_ly(
combined_pca_scores,
x = ~ PC1,
y = ~ PC2,
z = ~ PC3,
type = "scatter3d",
mode = "markers",
color = ~ stim_ordered
)18.2.2 PCA groupwise
# ------------------------------------------------------------------------------
# data formatting
# ------------------------------------------------------------------------------
# Convert the dataframe to wide format
df_wide.group <- pivot_wider(
subjectwise,
id_cols = c("tr_ordered", "stim_ordered"),
names_from = "sub",
values_from = "mean_per_sub"
)
# Split the data into two subsets based on the 'stim_ordered' value
# One for 'stimH' and another for 'stimL'
stim_high.df <- df_wide[df_wide$stim_ordered == "stimH",]
stim_low.df <- df_wide[df_wide$stim_ordered == "stimL",]
# Prepare data for PCA analysis by selecting relevant columns
# Exclude the first two columns and the last column
meanhighdf <-
data.frame(subset(stim_high.df, select = 3:(ncol(stim_high.df) - 1)))
meanlowdf <-
data.frame(subset(stim_low.df, select = 3:(ncol(stim_low.df) - 1)))
# ------------------------------------------------------------------------------
# Principal Component Analysis (PCA)
# ------------------------------------------------------------------------------
high.pca <- prcomp(meanhighdf) # Perform Principal Component Analysis (PCA)
high.pca_scores <- as.data.frame(high.pca$x) # Extract PCA scores
# Repeat the process for the low stimulus data
low.pca <- prcomp(meanlowdf)
low.pca_scores <- as.data.frame(low.pca$x)
combined_pca_scores <- rbind(high.pca_scores, low.pca_scores)
# Add a new column to indicate the 'stim_ordered' category (high_stim or low_stim)
# This helps in distinguishing the groups in the plot
combined_pca_scores$stim_ordered <-
c(rep("high_stim", nrow(high.pca_scores)), rep("low_stim", nrow(low.pca_scores)))
# ------------------------------------------------------------------------------
# plot 3D scatter plot of the PCA scores
# ------------------------------------------------------------------------------
# The points are colored based on their stim_ordered category
plot_ly(
combined_pca_scores,
x = ~ PC1,
y = ~ PC2,
z = ~ PC3,
type = "scatter3d",
mode = "markers",
color = ~ stim_ordered
)## Warning in RColorBrewer::brewer.pal(N, "Set2"): minimal value for n is 3, returning requested palette with 3 different levels
## Warning in RColorBrewer::brewer.pal(N, "Set2"): minimal value for n is 3, returning requested palette with 3 different levels
# ------------------------------------------------------------------------------
# plot 2D group plot
# ------------------------------------------------------------------------------
# Create a 2D plot with smoothed lines for each stim_ordered group
combined_pca <- combined_pca_scores %>%
group_by(stim_ordered) %>%
mutate(group_index = row_number())
ggplot(combined_pca,
aes(
x = group_index,
y = PC1,
group = stim_ordered,
colour = stim_ordered
)) +
stat_smooth(
method = "loess",
span = 0.25,
se = TRUE,
aes(color = stim_ordered),
alpha = 0.3
) +
theme_bw()## `geom_smooth()` using formula = 'y ~ x'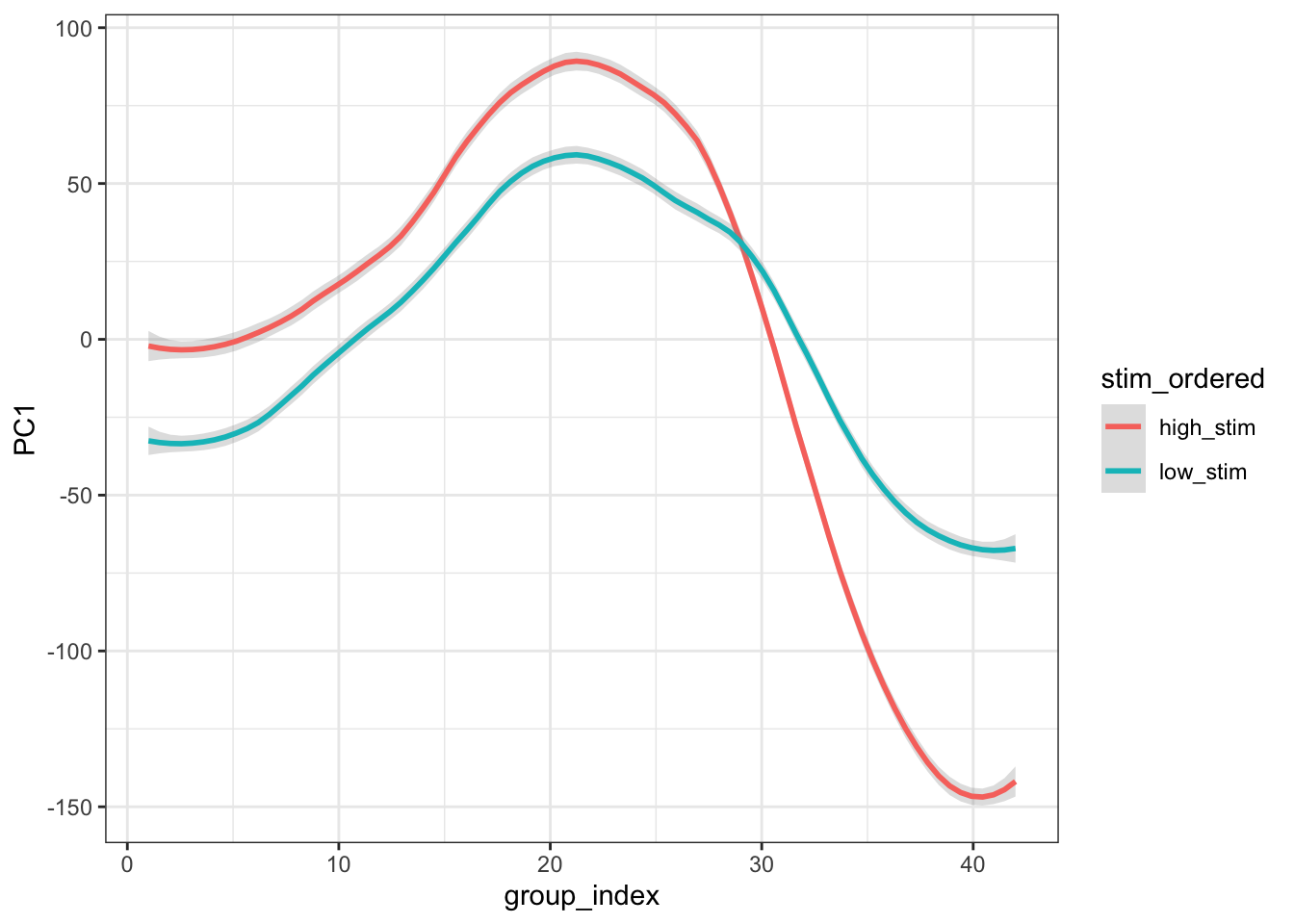
# Create the plot
ggplot(
groupwise,
aes(
x = tr_ordered,
y = mean_per_sub_mean,
group = stim_ordered,
colour = stim_ordered
)
) +
stat_smooth(
method = "loess",
span = 0.25,
se = TRUE,
aes(color = stim_ordered),
alpha = 0.3
) +
theme_bw()## `geom_smooth()` using formula = 'y ~ x'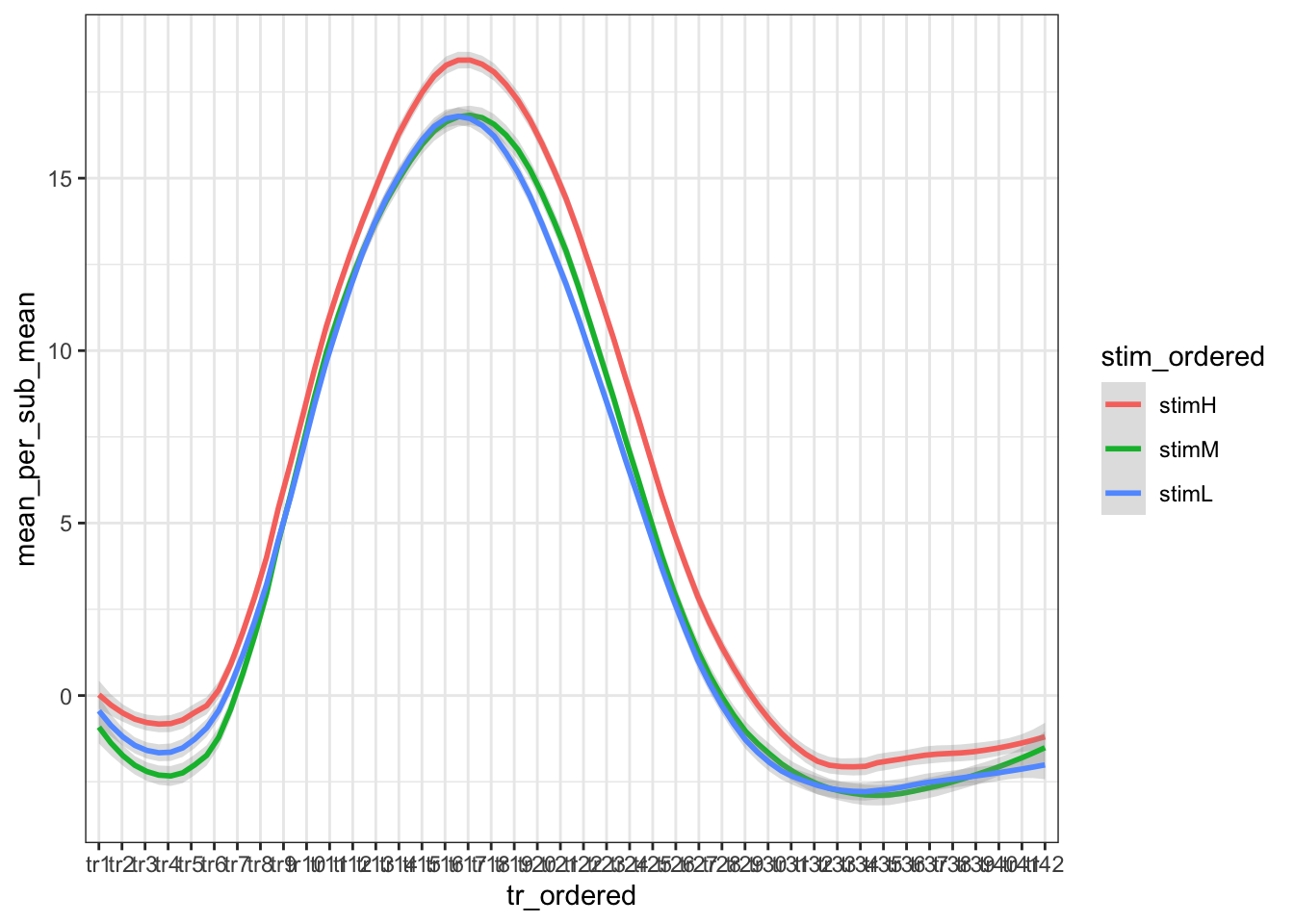
# Create the plot
# Create the plot with custom span and smoothing method
ggplot(groupwise, aes(x = tr_ordered, y = mean_per_sub_mean)) +
geom_line() + # Plot the smooth line for the mean
geom_ribbon(aes(ymin = mean_per_sub_mean - se, ymax = mean_per_sub_mean + se),
alpha = 0.3) + # Add the ribbon for standard error
geom_smooth(method = "loess", span = 0.1, se = FALSE) + # Add the loess smoothing curve
labs(x = "X-axis Label", y = "Y-axis Label", title = "Smooth Line with Standard Error Ribbon") +
theme_minimal()## `geom_smooth()` using formula = 'y ~ x'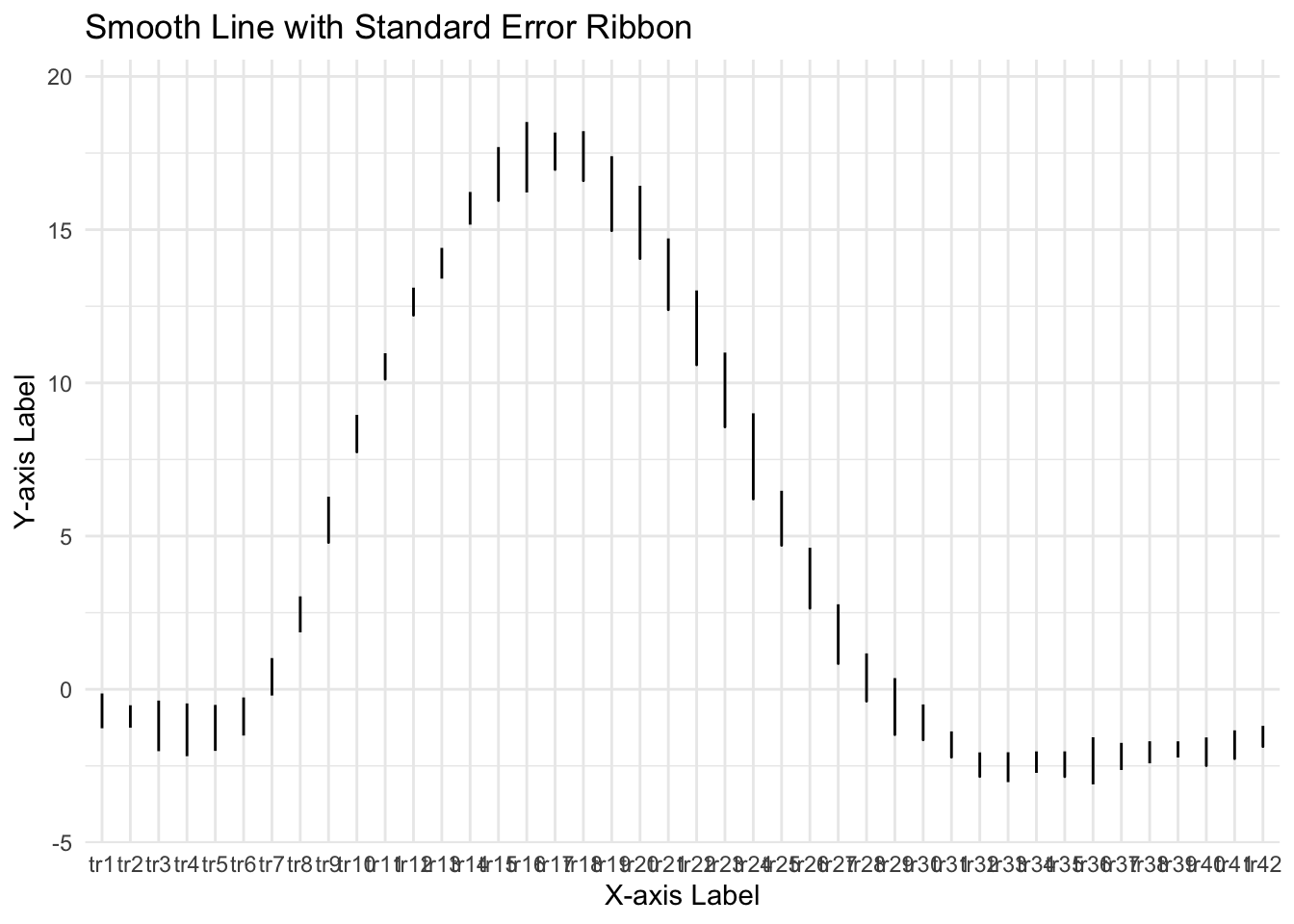
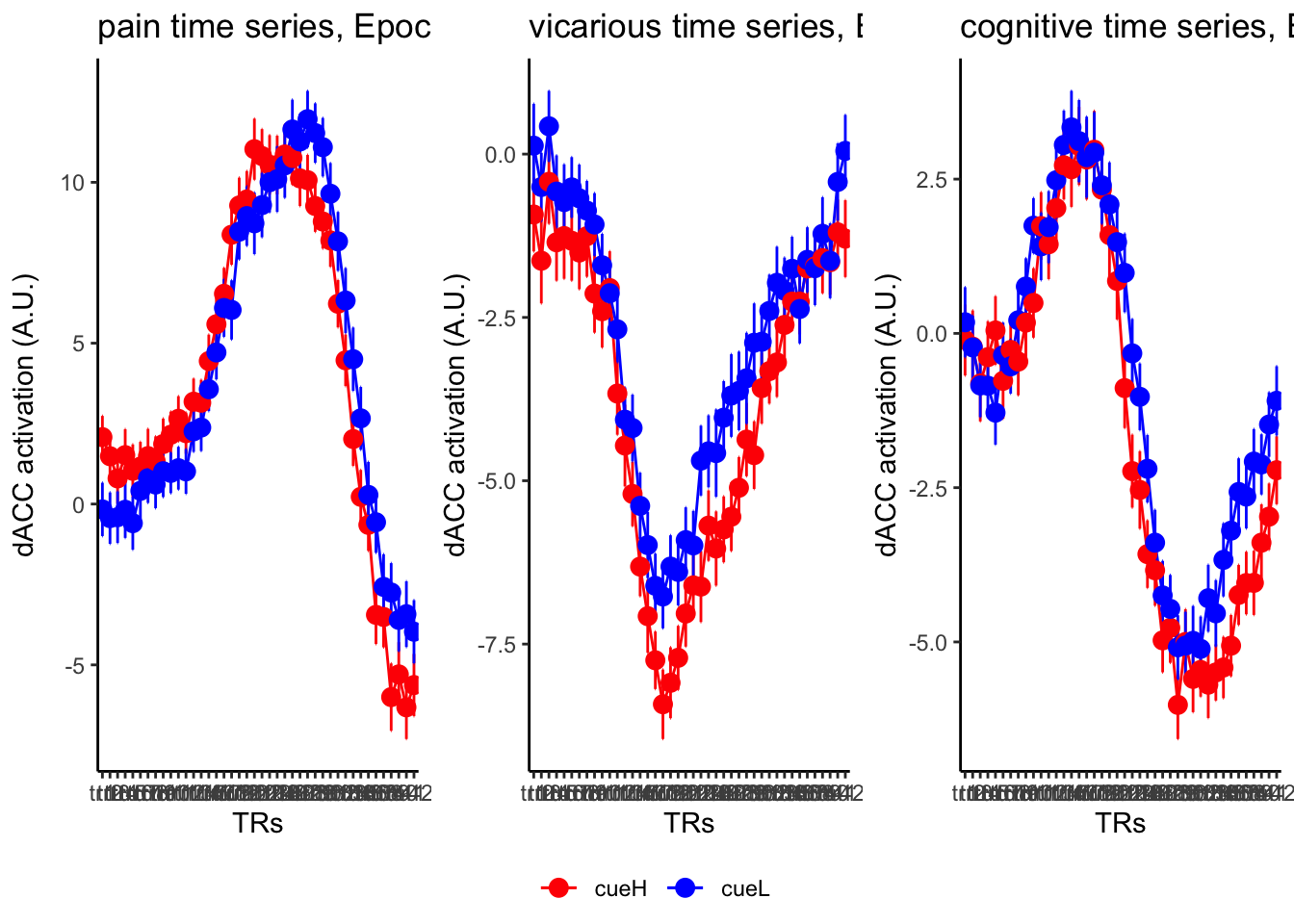
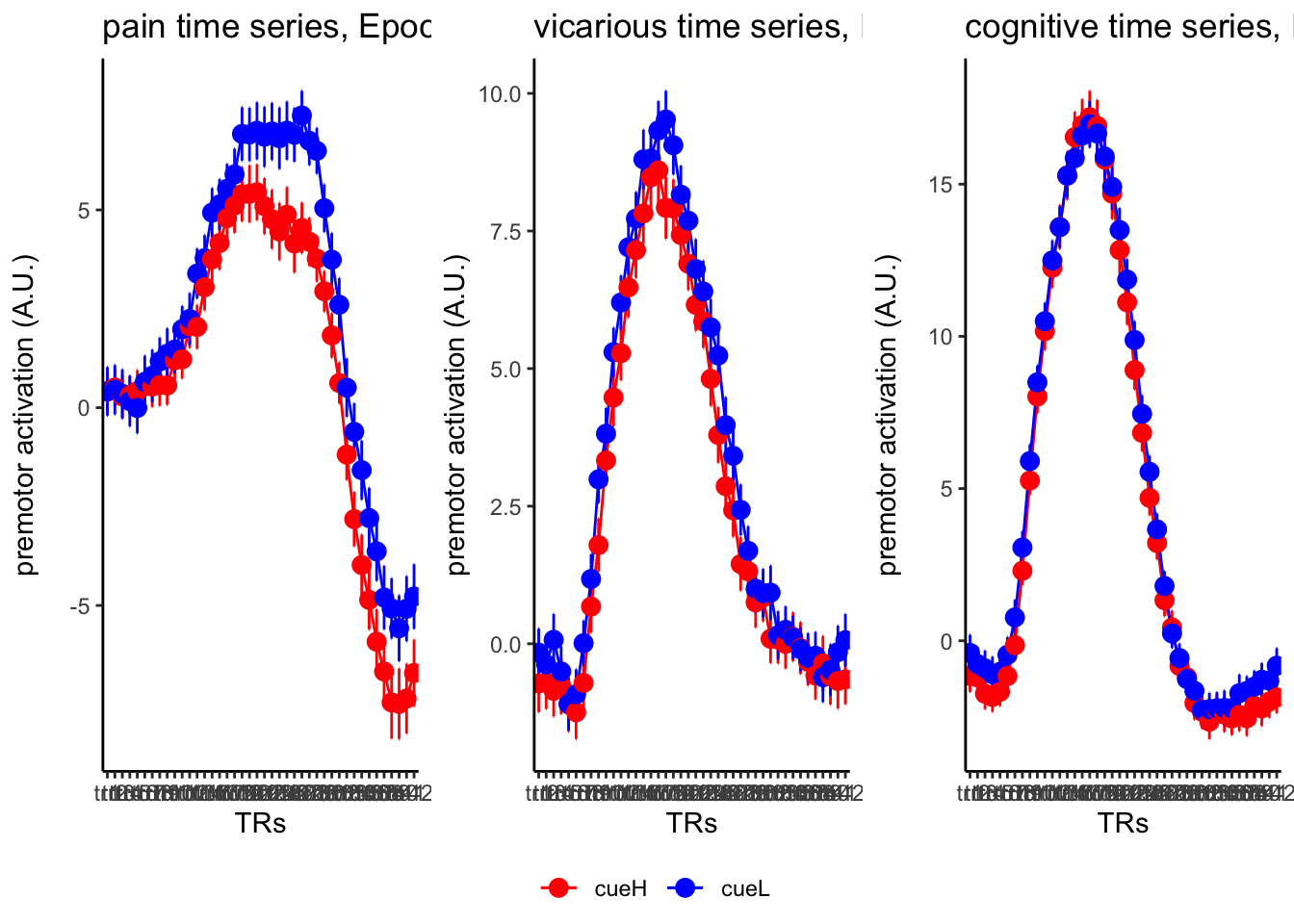
18.3 taskwise cue effect
roi_list <- c("dACC", "PHG", "V1", "SM", "MT", "RSC", "LOC", "FFC", "PIT", "pSTS", "AIP", "premotor") # 'rINS', 'TPJ',
for (ROI in roi_list) {
datadir <- file.path(main_dir, "analysis/fmri/spm/fir/ttl2par")
# taskname = 'pain'
exclude <- "sub-0001"
filename <- paste0("sub-*", "*roi-", ROI, "_tr-42.csv")
common_path <- Sys.glob(file.path(datadir, "sub-*", filename))
filter_path <-
common_path[!str_detect(common_path, pattern = exclude)]
df <-
do.call("rbind.fill", lapply(
filter_path,
FUN = function(files) {
read.table(files, header = TRUE, sep = ",")
}
))
run_types <- c("pain", "vicarious", "cognitive")
plot_list <- list()
TR_length <- 42
for (run_type in run_types) {
filtered_df <-
df[!(df$condition == "rating" |
df$condition == "cue" | df$runtype != run_type),]
parsed_df <- filtered_df %>%
separate(
condition,
into = c("cue", "stim"),
sep = "_",
remove = FALSE
)
# --------------------------------------------------------------------------
# subset regions based on ROI
# --------------------------------------------------------------------------
df_long <-
pivot_longer(
parsed_df,
cols = starts_with("tr"),
names_to = "tr_num",
values_to = "tr_value"
)
# --------------------------------------------------------------------------
# clean factor
# --------------------------------------------------------------------------
df_long$tr_ordered <- factor(df_long$tr_num,
levels = c(paste0("tr", 1:TR_length)))
df_long$cue_ordered <- factor(df_long$cue,
levels = c("cueH", "cueL"))
# --------------------------------------------------------------------------
# summary statistics
# --------------------------------------------------------------------------
subjectwise <- meanSummary(df_long,
c("sub", "tr_ordered", "cue_ordered"), "tr_value")
groupwise <- summarySEwithin(
data = subjectwise,
measurevar = "mean_per_sub",
withinvars = c("cue_ordered", "tr_ordered"),
idvar = "sub"
)
groupwise$task <- run_type
# https://stackoverflow.com/questions/29402528/append-data-frames-together-in-a-for-loop/29419402
LINEIV1 <- "tr_ordered"
LINEIV2 <- "cue_ordered"
MEAN <- "mean_per_sub_norm_mean"
ERROR <- "se"
dv_keyword <- "actual"
sorted_indices <- order(groupwise$tr_ordered)
groupwise_sorted <- groupwise[sorted_indices,]
p1 <- plot_timeseries_bar(
groupwise_sorted,
LINEIV1,
LINEIV2,
MEAN,
ERROR,
xlab = "TRs",
ylab = paste0(ROI, " activation (A.U.)"),
ggtitle = paste0(run_type, " time series, Epoch - stimulus"),
color = c("red", "blue")
)
time_points <- seq(1, 0.46 * TR_length, 0.46)
plot_list[[run_type]] <- p1 + theme_classic()
}
# ----------------------------------------------------------------------------
# plot three tasks
# ----------------------------------------------------------------------------
library(gridExtra)
plot_list <- lapply(plot_list, function(plot) {
plot + theme(plot.margin = margin(5, 5, 5, 5)) # Adjust plot margins if needed
})
combined_plot <-
ggpubr::ggarrange(
plot_list[["pain"]],
plot_list[["vicarious"]],
plot_list[["cognitive"]],
common.legend = TRUE,
legend = "bottom",
ncol = 3,
nrow = 1,
widths = c(3, 3, 3),
heights = c(.5, .5, .5),
align = "v"
)
print(combined_plot)
ggsave(file.path(
save_dir,
paste0("roi-", ROI, "_epoch-cue_desc-highcueGTlowcue.png")
),
combined_plot,
width = 12,
height = 4)
}
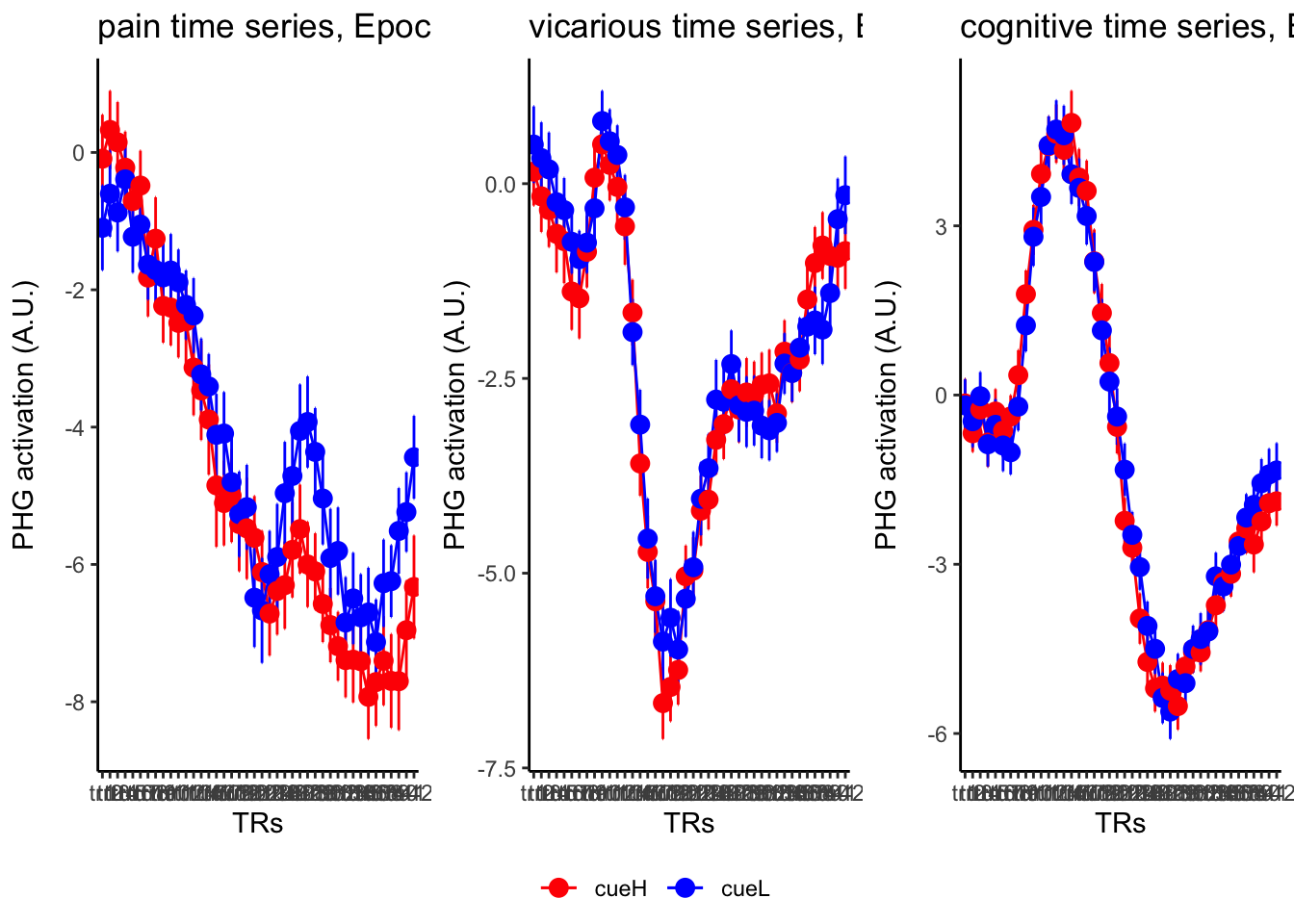
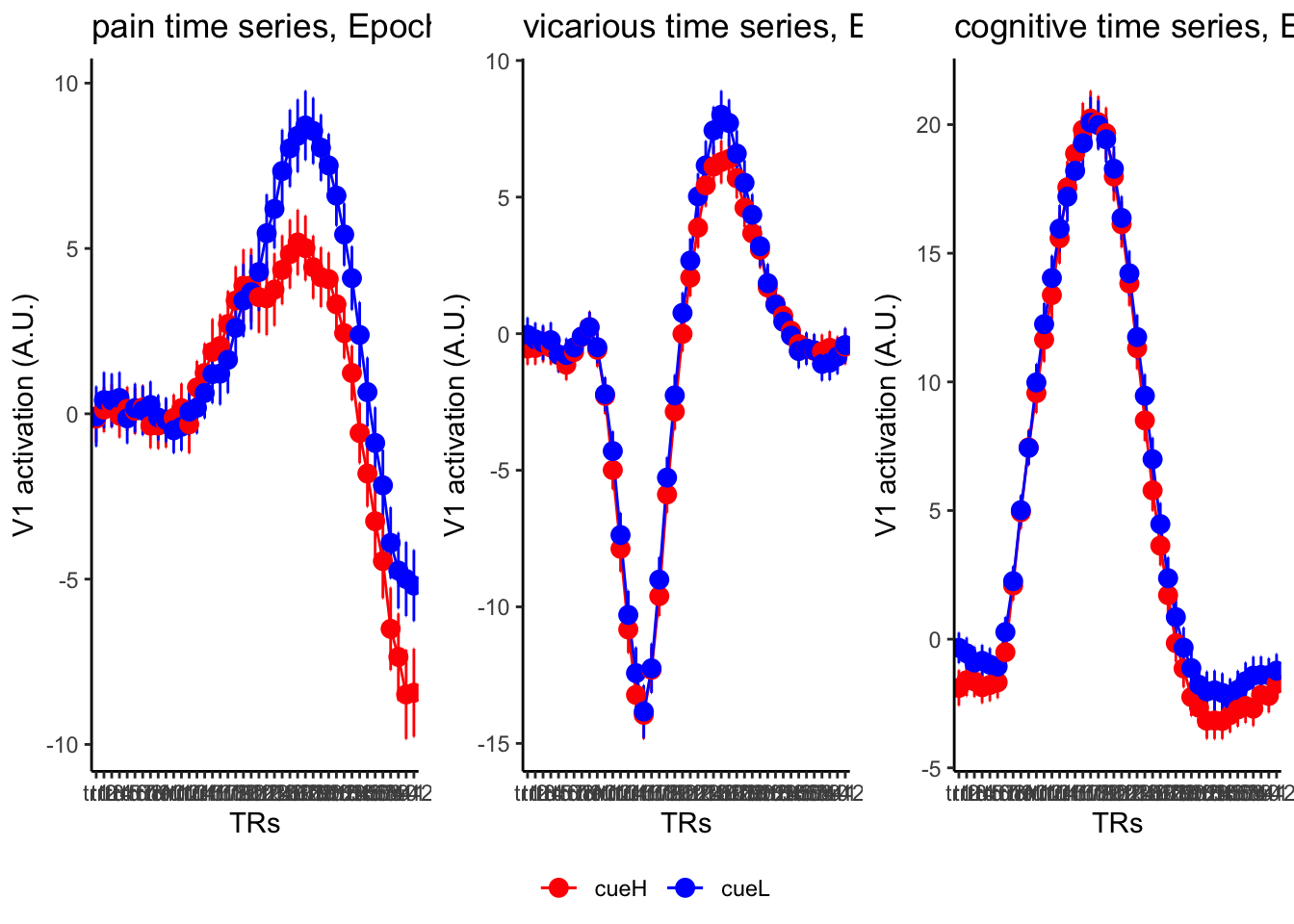
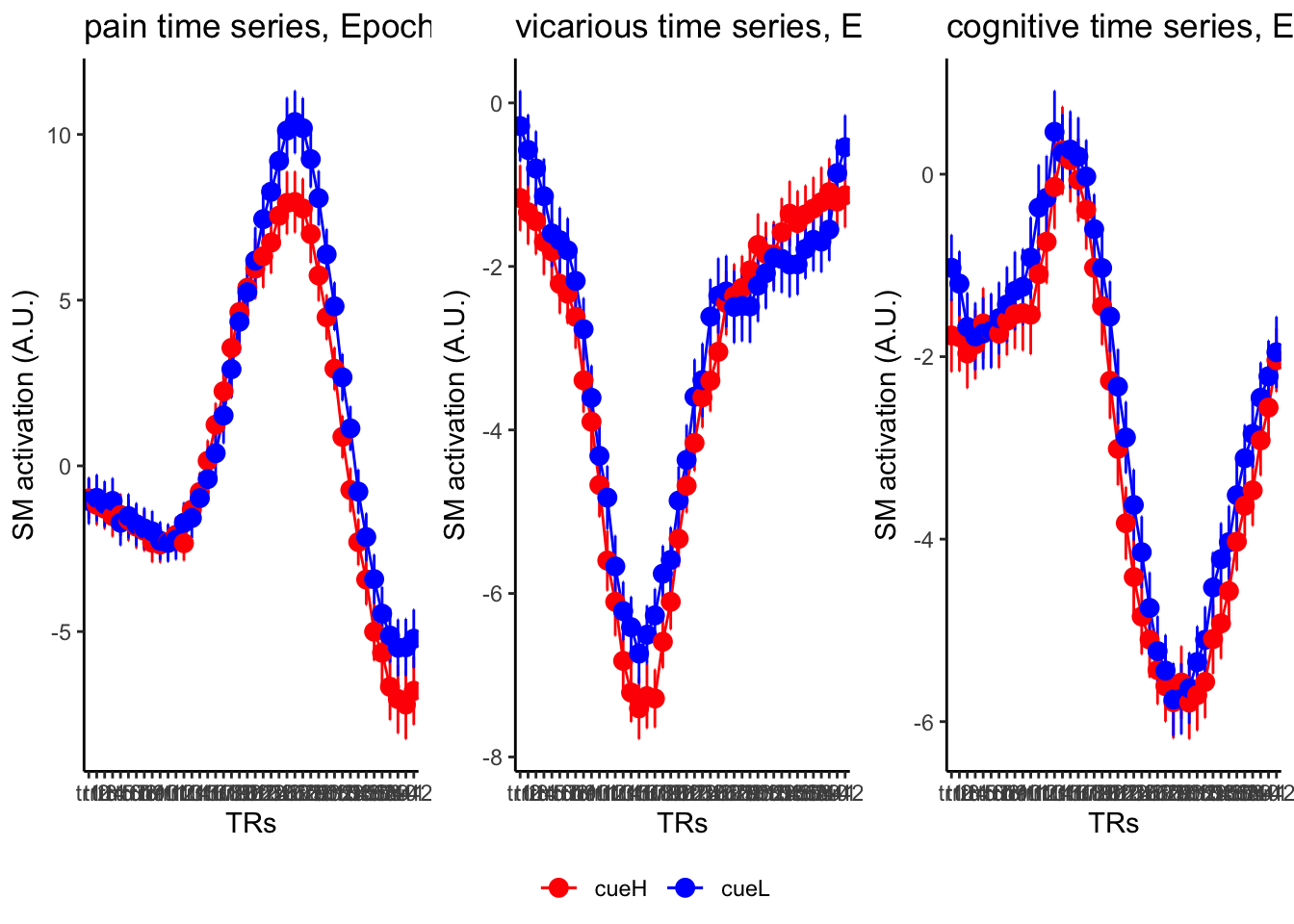
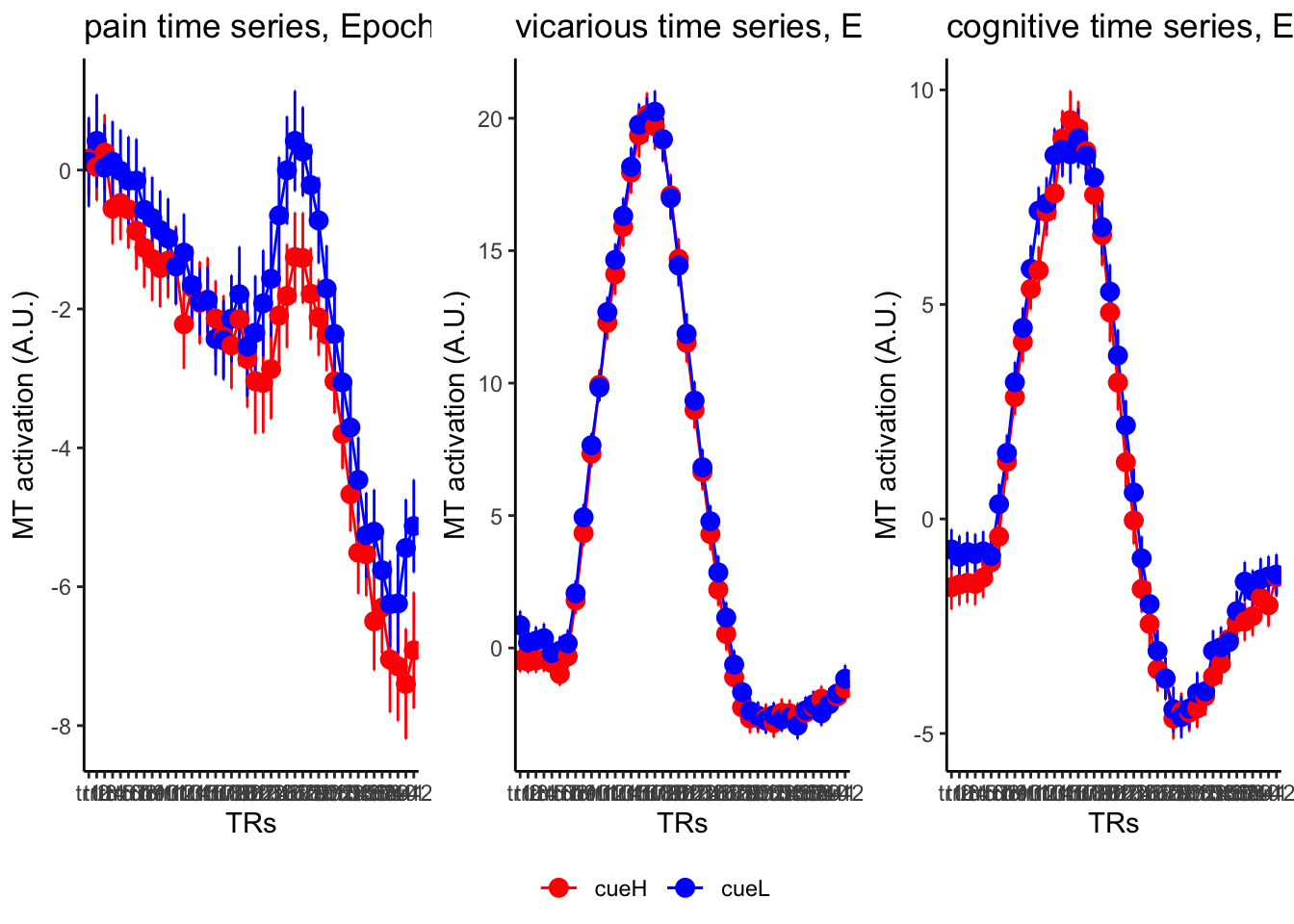
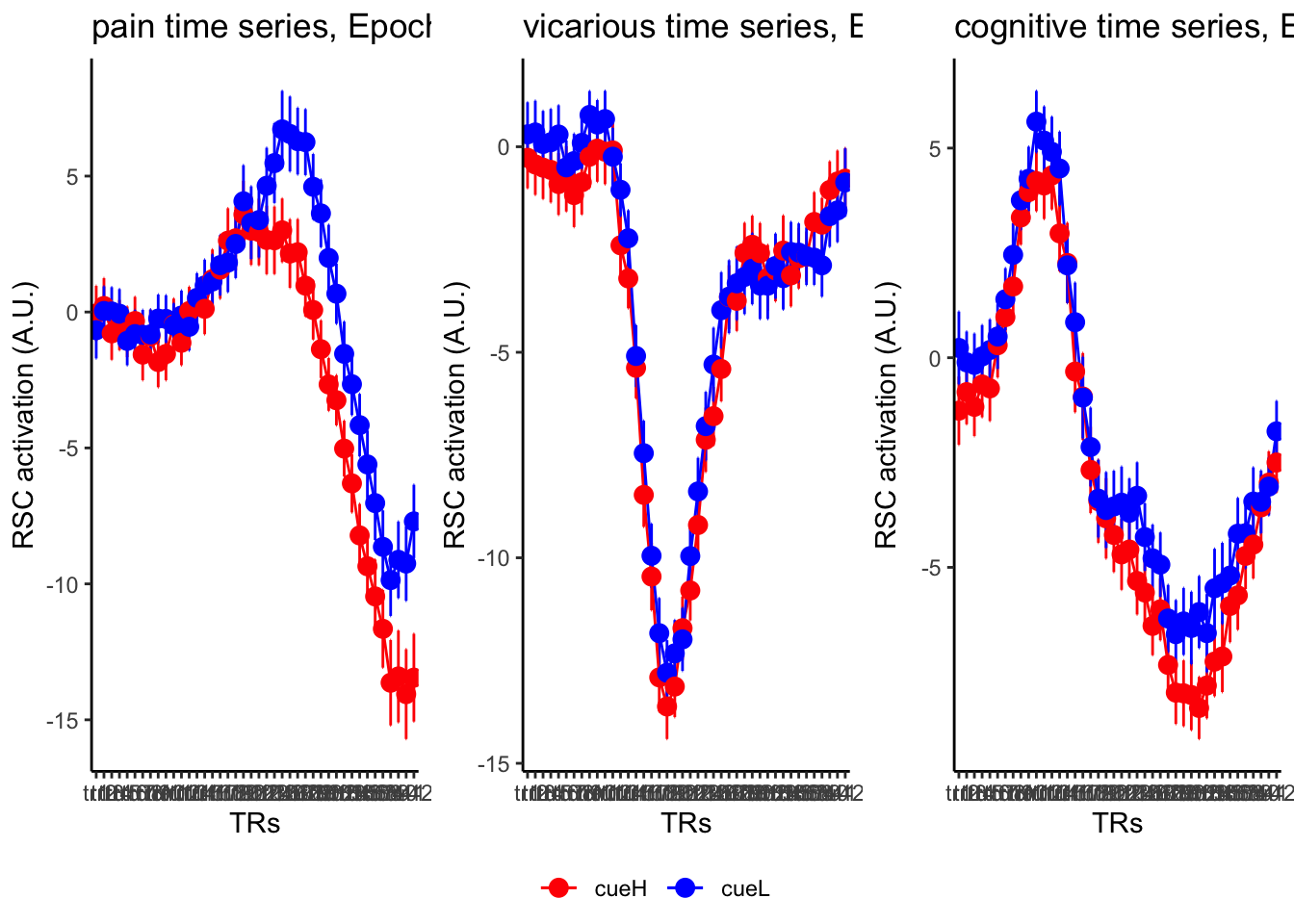
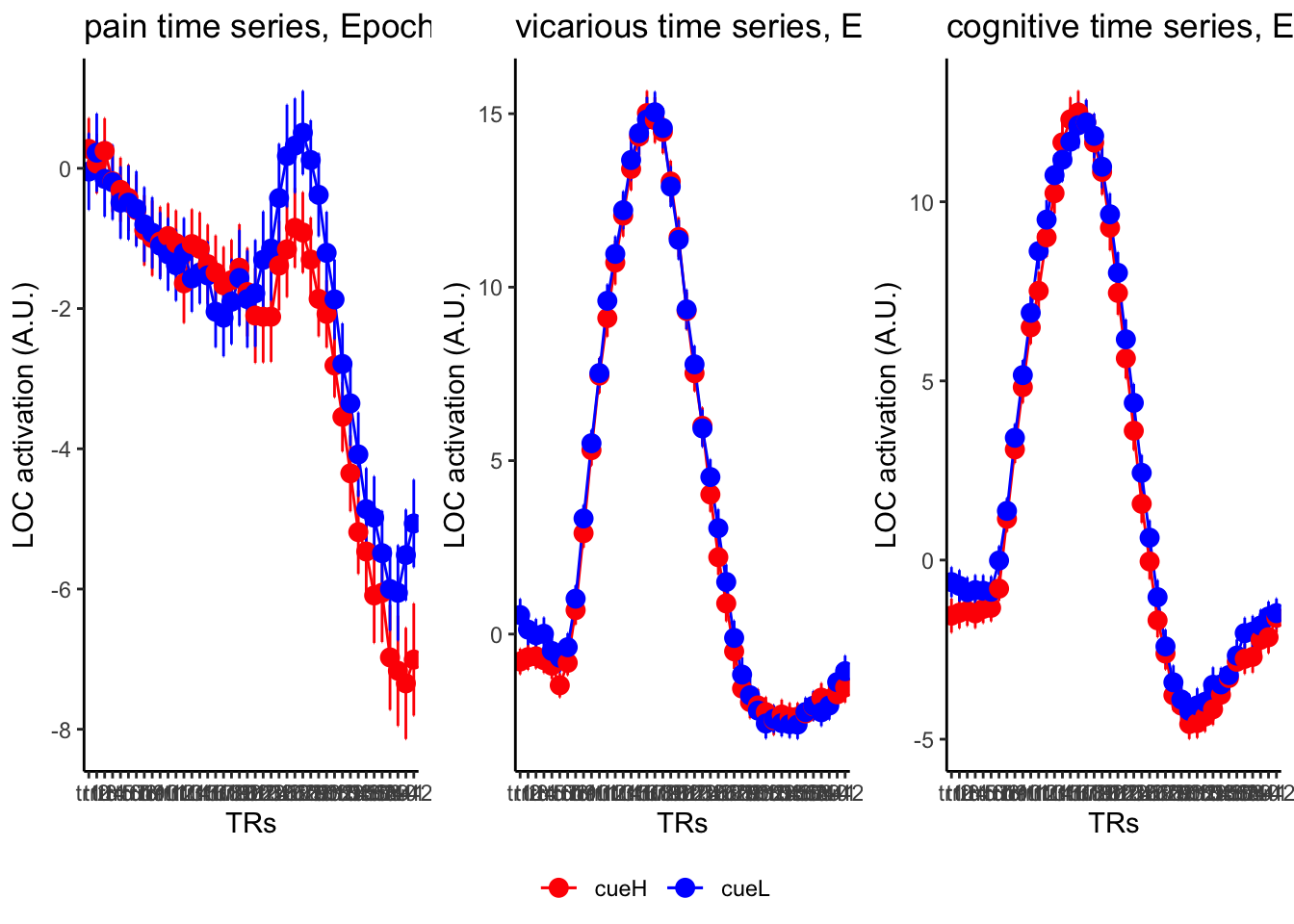
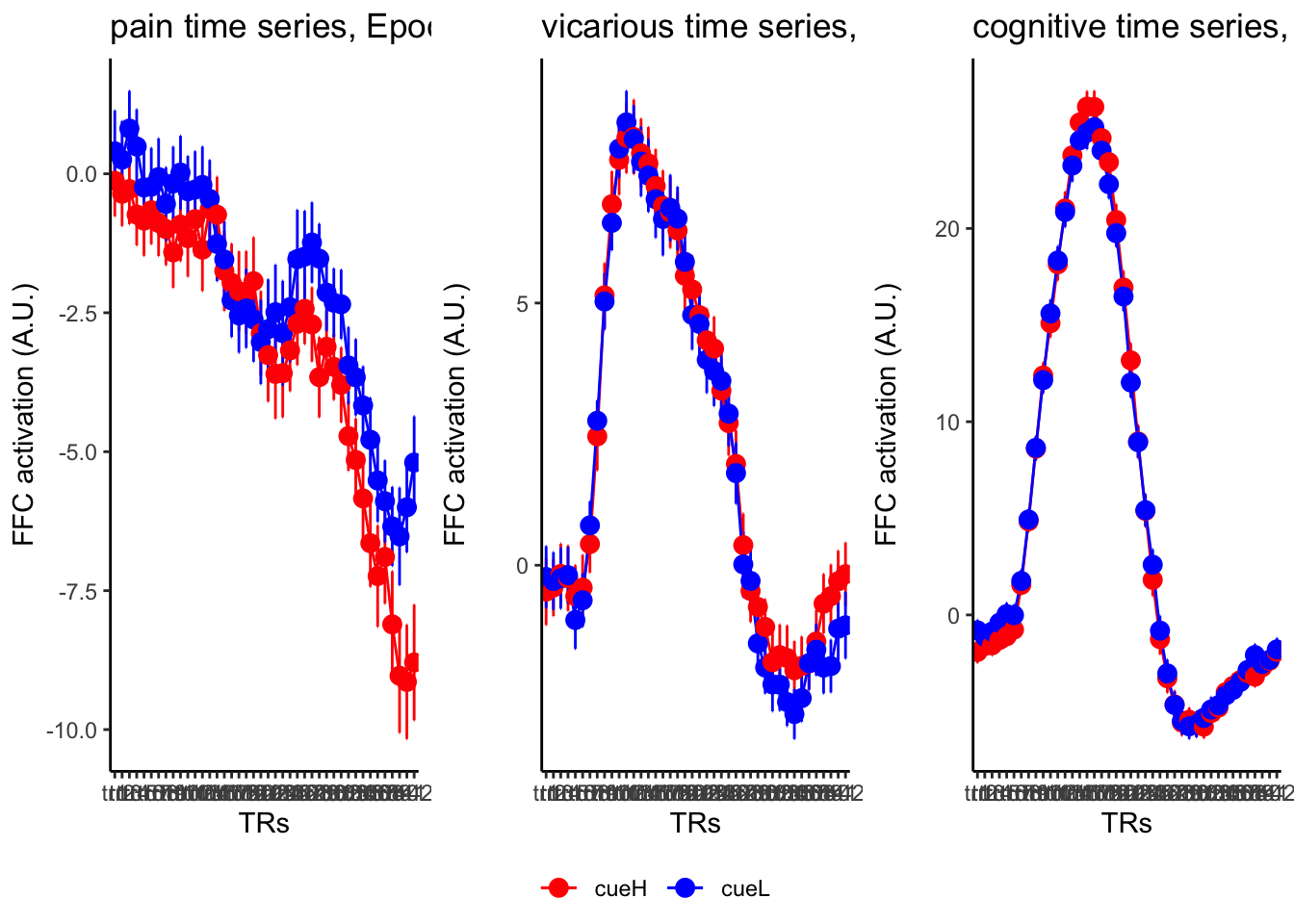
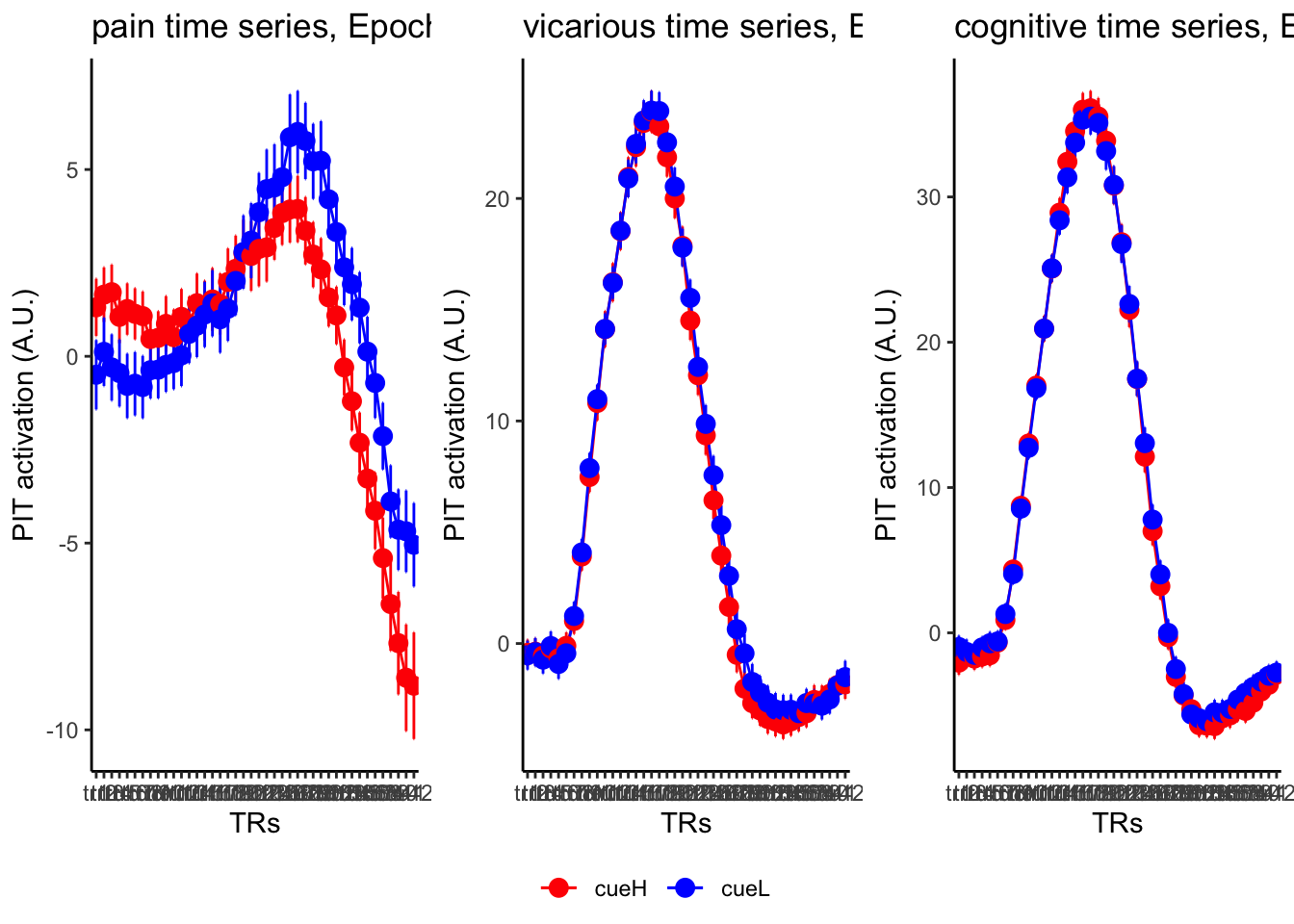
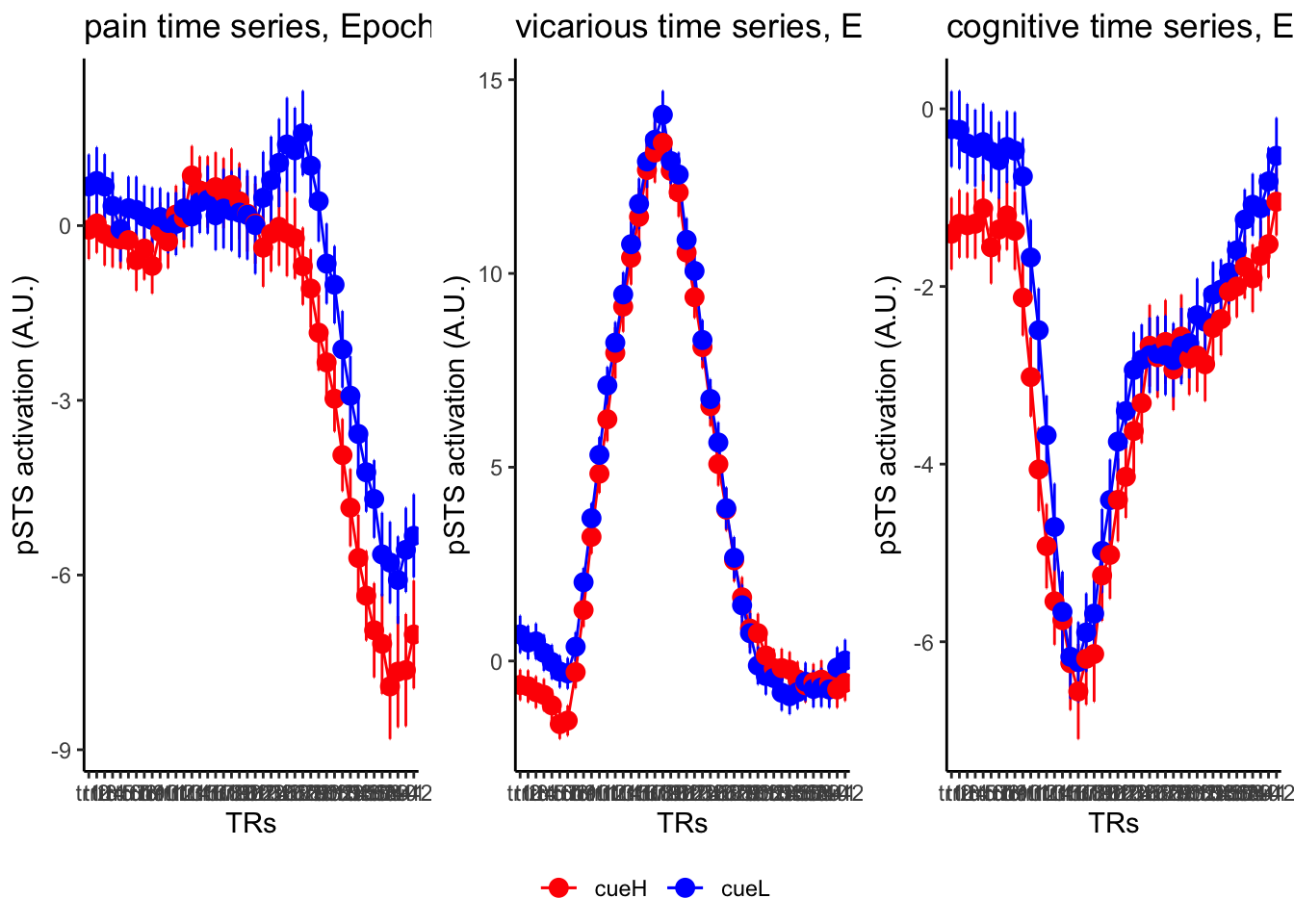
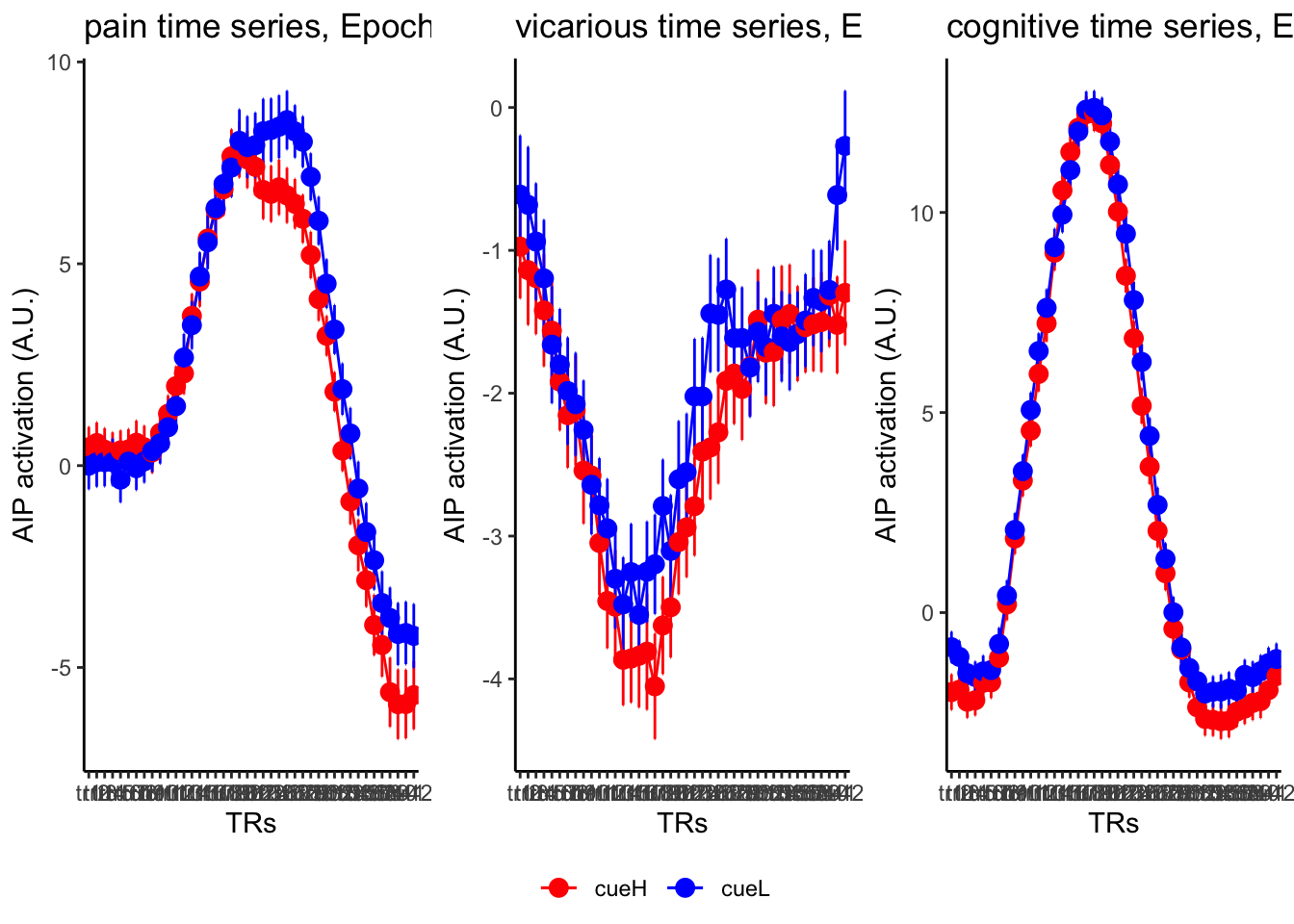
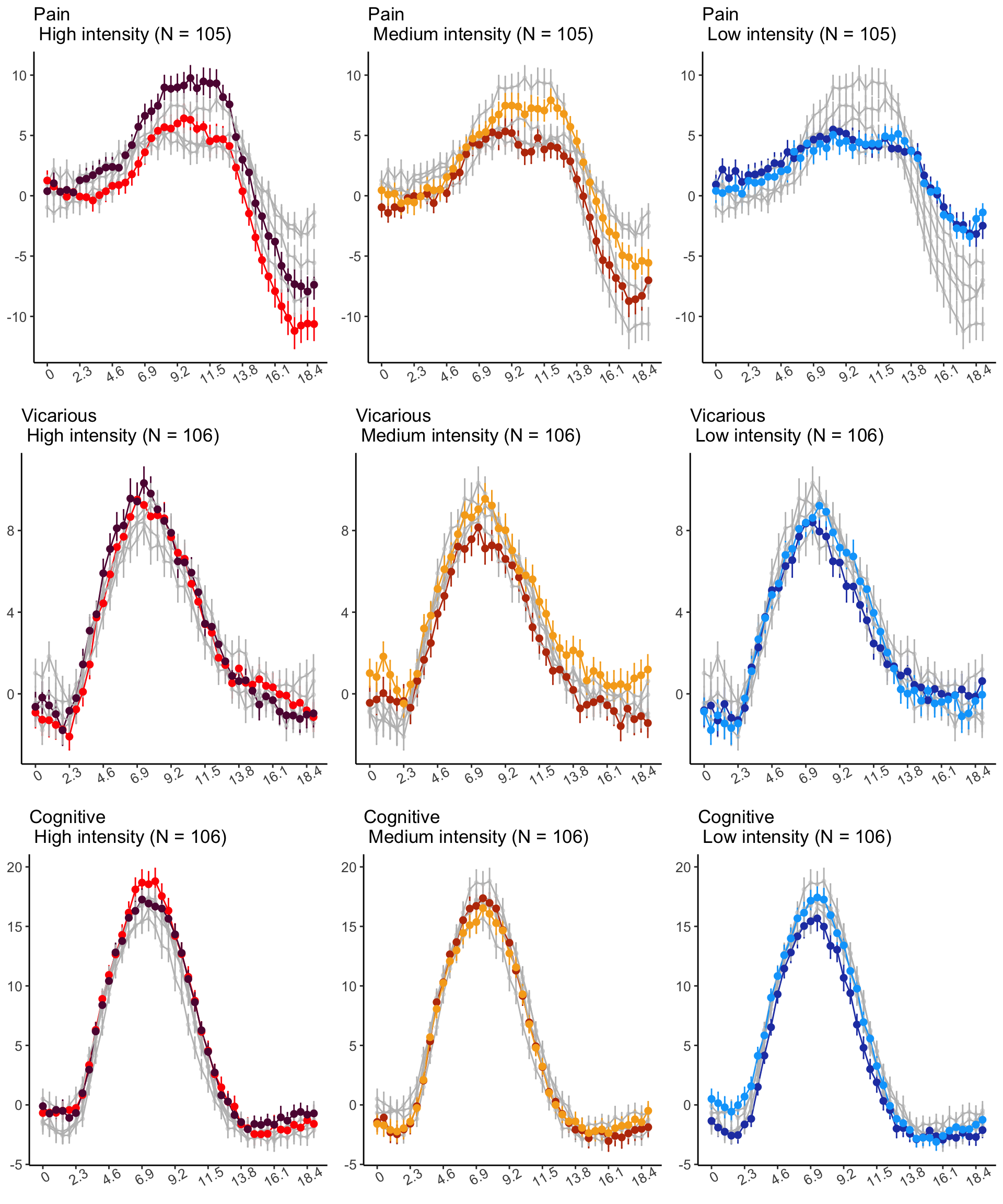
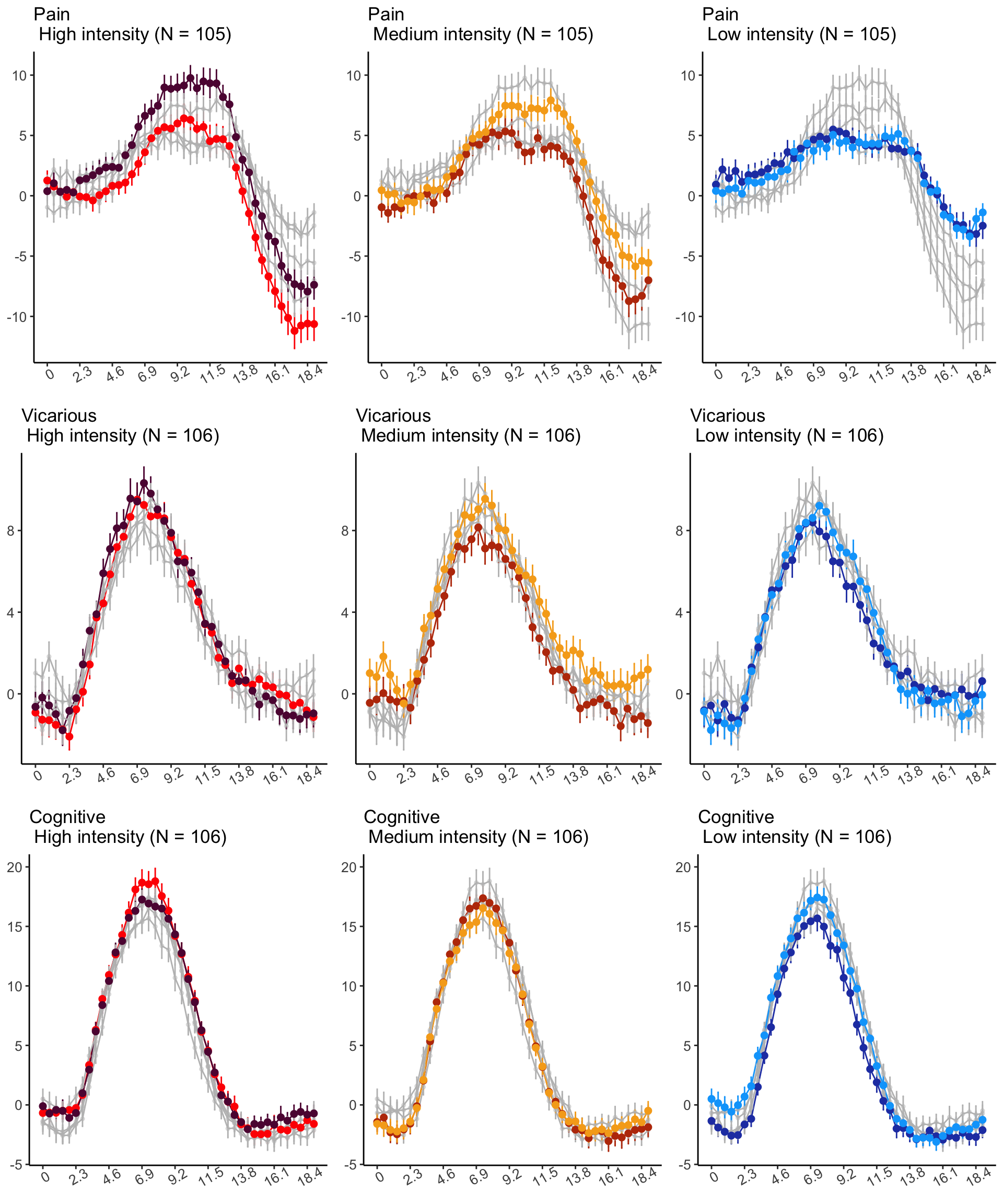
18.4 MAIN SANDBOX: 6 condition in three panels. per task. per ROI
# A function to plot data
plot_data <- function(groupwise, iv1, iv2, mean, error, xlab, ylab, ggtitle, run_type, colors) {
p <- plot_timeseries_bar(
groupwise,
"tr_ordered",
"sixcond",
"mean_per_sub_norm_mean",
"se",
xlab = "TRs",
ylab = "Epoch: stimulus, High cue vs. Low cue",
ggtitle = paste0(run_type, " intensity (N = ", unique(groupwise$N), ")"),
color_mapping = colors,
show_legend = FALSE
)
p + theme_classic()
}
#' Calculate Point Size Proportionally
#'
#' This function calculates point size proportionally based on a base point size and figure dimensions
#' (width and height). It can be used to adjust the point size in plots to maintain proportionality
#' with varying figure sizes.
#'
#' @param point_size_base The base point size for `geom_points`.
#' @param figure_width The width of the figure in which the point size needs to be adjusted.
#' @param figure_height The height of the figure in which the point size needs to be adjusted.
#'
#' @return The calculated point size.
#'
#' @examples
#' # Define your point size base
#' point_size_base <- 3
#'
#' # Define your figure dimensions (width and height)
#' figure_width <- 12
#' figure_height <- 8
#'
#' # Calculate the point size using the function
#' POINT_SIZE <- calculate_point_size(point_size_base, figure_width, figure_height)
#'
#' # Apply the point size to your plot elements
#' plot + geom_point(size = POINT_SIZE)
#'
#' @export
calculate_point_size <- function(figure_width, figure_height, point_size_base = 5) {
scaling_factor <- min(figure_width, figure_height) / point_size_base
return(scaling_factor)
}
# ------------------------------------------------------------------------------
# epoch stim, high cue vs low cue
# ------------------------------------------------------------------------------
run_types <- c("pain", "vicarious", "cognitive")
all_plots <- list()
TR_length <- 42
for (roi in c("dACC", "PHG")) {
plot_list_per_roi <- list()
for (run_type in run_types) {
filtered_df <-
df[!(
df$condition == "rating" |
df$condition == "cue" |
df$runtype != run_type | df$ROI == roi
),]
plot_list <- list()
parsed_df <- filtered_df %>%
separate(
condition,
into = c("cue", "stim"),
sep = "_",
remove = FALSE
)
# --------------------- subset regions based on ROI ----------------------------
df_long <-
pivot_longer(
parsed_df,
cols = starts_with("tr"),
names_to = "tr_num",
values_to = "tr_value"
)
# ----------------------------------------------------------------------------
# clean factor
# ----------------------------------------------------------------------------
df_long$tr_ordered <- factor(df_long$tr_num,
levels = c(paste0("tr", 1:TR_length)))
df_long$cue_ordered <- factor(df_long$cue,
levels = c("cueH", "cueL"))
df_long$stim_ordered <- factor(df_long$stim,
levels = c("stimH", "stimM", "stimL"))
df_long$sixcond <- factor(
df_long$condition,
levels = c(
"cueH_stimH",
"cueL_stimH",
"cueH_stimM",
"cueL_stimM",
"cueH_stimL",
"cueL_stimL"
)
)
# ------------------------------------------------------------------------------
# summary statistics
# ------------------------------------------------------------------------------
subjectwise <- meanSummary(df_long,
c("sub", "tr_ordered", "sixcond"), "tr_value")
groupwise <- summarySEwithin(
data = subjectwise,
measurevar = "mean_per_sub",
withinvars = c("sixcond", "tr_ordered"),
idvar = "sub"
)
groupwise$task <- taskname
# ----------------------------------------------------------------------------
# plot parameters
# ----------------------------------------------------------------------------
# convert TR orders to numeric values
tr_numbers <-
as.numeric(sub("tr", "", as.character(groupwise$tr_ordered)))
tr_sequence <- (tr_numbers - 1) * 0.46
groupwise$tr_sequence <- tr_sequence
LINEIV1 <- "tr_sequence"
LINEIV2 <- "sixcond"
MEAN <- "mean_per_sub_norm_mean"
ERROR <- "se"
dv_keyword <- "actual"
sorted_indices <- order(groupwise$tr_ordered)
groupwise_sorted <- groupwise[sorted_indices,]
XLAB <- "TRs"
YLAB <- "Stimulus Epoch High vs. Low cue"
HIGHSTIM_COLOR <- c(
"cueH_stimH" = "red",
"cueL_stimH" = "#5f0f40",
"cueH_stimM" = "gray",
"cueL_stimM" = "gray",
"cueH_stimL" = "gray",
"cueL_stimL" = "gray"
)
MEDSTIM_COLOR <- c(
"cueH_stimH" = "gray",
"cueL_stimH" = "gray",
"cueH_stimM" = "#bc3908",
"cueL_stimM" = "#f6aa1c",
"cueH_stimL" = "gray",
"cueL_stimL" = "gray"
)
LOWSTIM_COLOR <- c(
"cueH_stimH" = "gray",
"cueL_stimH" = "gray",
"cueH_stimM" = "gray",
"cueL_stimM" = "gray",
"cueH_stimL" = "#2541b2",
"cueL_stimL" = "#00a6fb"
)
AXIS_FONTSIZE <- 10
COMMONAXIS_FONTSIZE <- 15
TITLE_FONTSIZE <- 20
figure_width <- 10 # Adjust this to your actual figure width
figure_height <- 10 # Adjust this to your actual figure height
GEOMPOINT_SIZE <- calculate_point_size(figure_width, figure_height)
# ----------------------------------------------------------------------------
# plot intensity per task
# ----------------------------------------------------------------------------
p3H <- plot_timeseries_bar(
groupwise,
LINEIV1,
LINEIV2,
MEAN,
ERROR,
XLAB,
YLAB,
ggtitle = paste0(tools::toTitleCase(run_type), "\n High intensity (N = ", unique(groupwise$N), ")"),
color_mapping = HIGHSTIM_COLOR,
show_legend = FALSE,
geompoint_size = GEOMPOINT_SIZE
)
# Assuming tr_sequence is correct and has been added to groupwise
# Calculate breaks to show every 10th TR
breaks_to_show <-
seq(0, max(groupwise$tr_sequence), by = 0.46 * 5)
labels_to_show <-
seq(0, max(groupwise$tr_sequence), by = 0.46 * 5)
# It's important to ensure that both 'breaks_to_show' and 'labels_to_show' have the same length
# If the lengths differ, we need to adjust them so they match
if (length(breaks_to_show) != length(labels_to_show)) {
# Assuming you want to keep all the breaks and just adjust the labels
labels_to_show <- labels_to_show[seq_along(breaks_to_show)]
}
# High intensity
plot_list[["H"]] <- p3H +
scale_x_continuous(
breaks = breaks_to_show,
# Set breaks at every 10th point
labels = labels_to_show,
# Use the calculated labels
limits = range(groupwise$tr_sequence) # Set the limits based on the sequence
) +
theme_classic()
# Medium intensity
p3M <- plot_timeseries_bar(
groupwise,
LINEIV1,
LINEIV2,
MEAN,
ERROR,
XLAB,
YLAB,
ggtitle = paste0(
tools::toTitleCase(run_type),
"\n Medium intensity (N = ",
unique(groupwise$N),
")"
),
color_mapping = MEDSTIM_COLOR,
show_legend = FALSE,
geompoint_size = GEOMPOINT_SIZE
)
plot_list[["M"]] <- p3M +
scale_x_continuous(
breaks = breaks_to_show, # Set breaks at every 10th point
labels = labels_to_show, # Use the calculated labels
limits = range(groupwise$tr_sequence) # Set the limits based on the sequence
) +
theme_classic()
# Low intensity
p3L <- plot_timeseries_bar(
groupwise,
LINEIV1,
LINEIV2,
MEAN,
ERROR,
XLAB,
YLAB,
ggtitle = paste0(tools::toTitleCase(run_type), "\n Low intensity (N = ", unique(groupwise$N), ")"),
color_mapping = LOWSTIM_COLOR,
show_legend = FALSE,
geompoint_size = GEOMPOINT_SIZE
)
plot_list[["L"]] <- p3L +
scale_x_continuous(
breaks = breaks_to_show,
# Set breaks at every 10th point
labels = labels_to_show,
# Use the calculated labels
limits = range(groupwise$tr_sequence) # Set the limits based on the sequence
) +
theme_classic()
# ----------------------------------------------------------------------------
# combine three tasks in one panel per ROI
# ----------------------------------------------------------------------------
library(gridExtra)
plot_list <- lapply(plot_list, function(plot) {
plot +
theme(
plot.margin = margin(5, 5, 5, 5), # Adjust plot margins if needed
axis.title.y = element_blank(), # Remove y-axis title
axis.title.x = element_blank(),
axis.text.y = element_text(size = AXIS_FONTSIZE), # Increase y-axis text size
axis.text.x = element_text(size = AXIS_FONTSIZE, angle = 30)
)
})
combined_plot_per_run <-
ggpubr::ggarrange(
plot_list[["H"]],
plot_list[["M"]],
plot_list[["L"]],
common.legend = FALSE,
legend = "none",
ncol = 3,
nrow = 1,
widths = c(3, 3, 3),
heights = c(.5, .5, .5),
align = "v"
)
# Add the combined plot for this run type to the list for the current ROI
plot_list_per_roi[[run_type]] <- combined_plot_per_run
} # end of run loop
# ----------------------------------------------------------------------------
# add commom legend
# ----------------------------------------------------------------------------
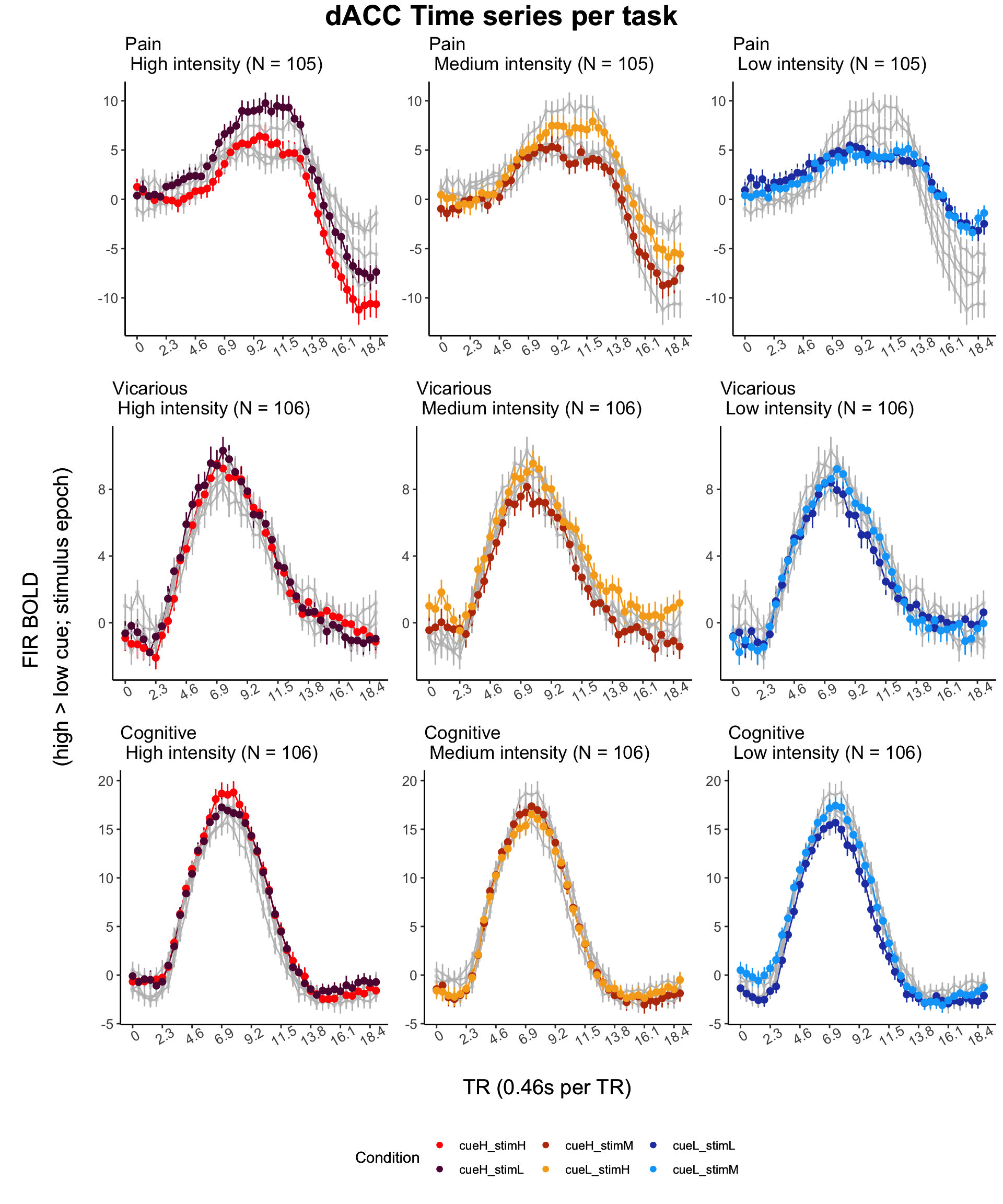
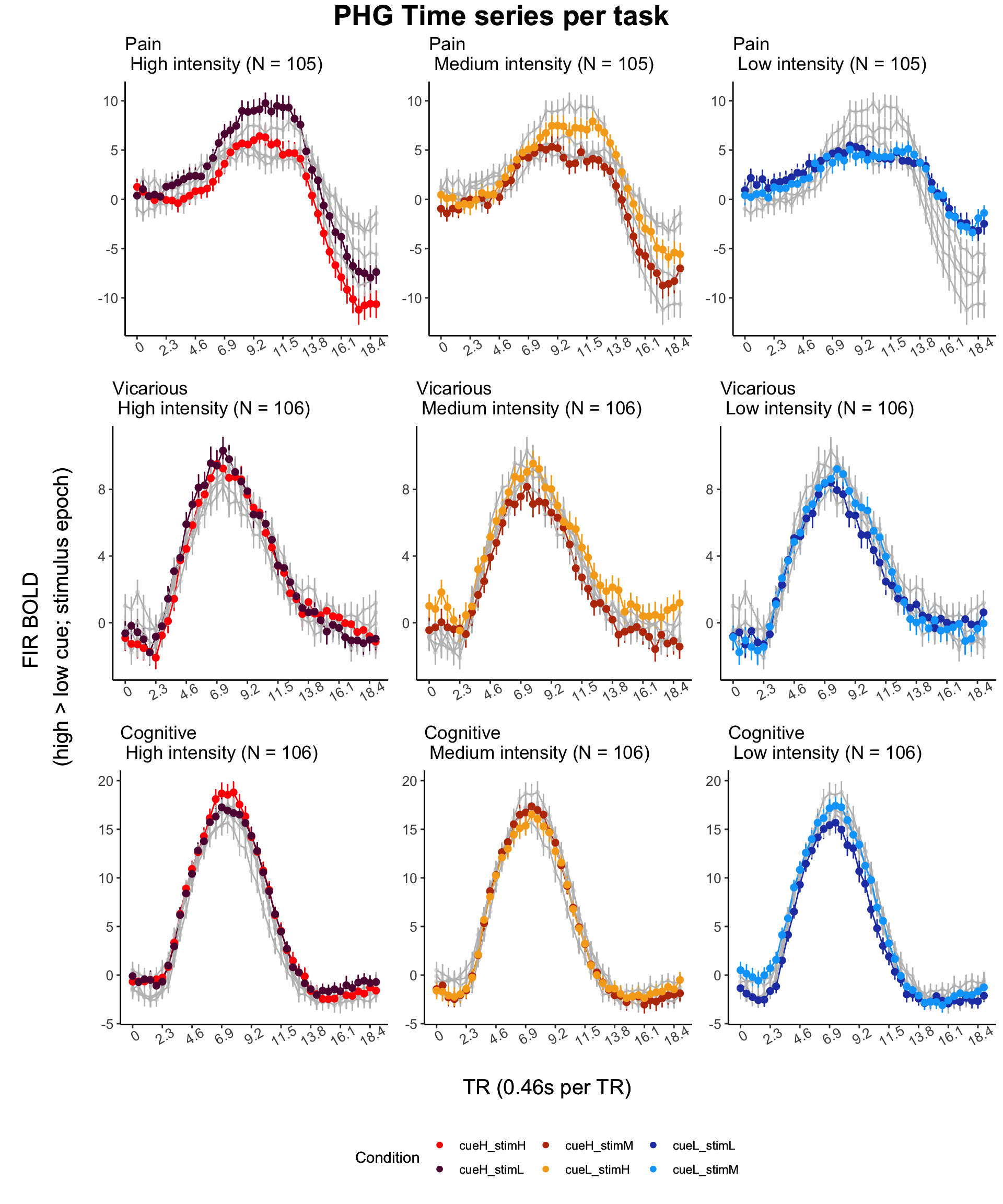
legend_data <- data.frame(
sixcond = factor(
c(
"cueH_stimH",
"cueL_stimH",
"cueH_stimM",
"cueL_stimM",
"cueH_stimL",
"cueL_stimL"
)
),
color = c("red", "#5f0f40", "#bc3908", "#f6aa1c", "#2541b2", "#00a6fb"),
stringsAsFactors = FALSE
)
legend_plot <-
ggplot(legend_data, aes(x = sixcond, y = 1, color = sixcond)) +
geom_point() +
scale_color_manual(values = legend_data$color) +
theme_void() +
theme(legend.position = "bottom") +
guides(color = guide_legend(title = "Condition"))
legend_grob <-
ggplotGrob(legend_plot)$grobs[[which(sapply(ggplotGrob(legend_plot)$grobs, function(x)
x$name) == "guide-box")]]
heights <- c(rep(1, length(run_types)), 2)
# ----------------------------------------------------------------------------
# common axes for the 9 panels
# ----------------------------------------------------------------------------
y_axis_label <-
textGrob(
"FIR BOLD \n(high > low cue; stimulus epoch)",
rot = 90,
gp = gpar(fontsize = COMMONAXIS_FONTSIZE)
)
x_axis_label <-
textGrob(
"TR (0.46s per TR)",
rot = 0,
gp = gpar(fontsize = COMMONAXIS_FONTSIZE)
)
num_rows <- length(plot_list_per_roi) + 1 # +1 for the legend
# ----------------------------------------------------------------------------
# combined plots across 3 tasks
# ----------------------------------------------------------------------------
roi_combined_plot <-
do.call(grid.arrange, c(plot_list_per_roi, ncol = 1))
final_plot <- grid.arrange(
y_axis_label,
arrangeGrob(
roi_combined_plot,
x_axis_label,
legend_grob,
ncol = 1,
heights = c(11,.5, 1)
),
ncol = 2,
widths = c(1, 10), # Relative widths for the label, plots, and legend,
top = textGrob(sprintf("%s Time series per task", roi), gp = gpar(
fontsize = TITLE_FONTSIZE, fontface = "bold"
)) # title parameter
)
grid.draw(final_plot)
# ----------------------------------------------------------------------------
# save all plots
# ----------------------------------------------------------------------------
ggsave(file.path(
save_dir,
paste0("roi-",
roi ,
"_epoch-stim_desc-stimcuecomparison.png")
),
all_plots[[roi]],
width = 12,
height = 20)
}